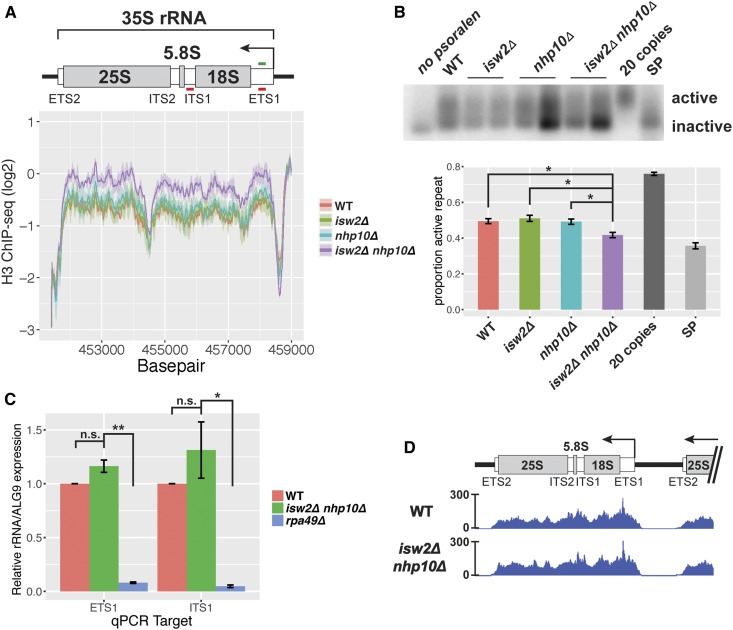
Figure 2.
Nucleosome occupancy, but not transcription, is affected at the 35S rDNA in isw2∆ and nhp10∆ mutants. (A) Histone H3 ChIP-seq through the 35S rRNA gene. Line represents average log2 ChIP-seq signal at each base pair for two independent experiments, and the ribbon represents the SEM at each base pair. Schematic drawing of the 35S indicates transcribed spacers that are removed during processing, as well as the mature 18S, 5.8S, and 25S rRNAs that are parts of complete ribosomes. ETS1 and ITS1 qPCR primer sets are indicated with red lines, and ETS1 hybridization probe, used in the Southern blot shown in (B), indicated in green. (B) Psoralen cross-linked DNA, digested with EcoRI and hybridized with a probe to the ETS1 region. Two independent isolates of each remodeling factor mutant are shown. For quantification, signal strengths of the active and inactive bands were measured with ImageJ software, and the proportion of the total signal present in the “active” band was calculated. Values for each genotype reflect between three and five biological replicates, and error bars represent SEM. The “20 copies” sample comes from a strain with only 20 copies of the rDNA repeat, in which all 20 copies should be actively transcribed, and “SP” is the wild-type (WT) strain at stationary phase, when a large proportion of rDNA repeats should be inactive. (C) RT-qPCR measuring the ETS1 and ITS1 of the 35S pre-rRNA. For each qPCR target, expression for all strains normalized to WT. For (B) and (C), statistical significance determined by pairwise t-tests followed by Bonferroni correction for multiple testing. * P < 0.05, ** P < 0.005. (D) RNA Pol I ChIP-seq (FLAG-tagged RNA Pol I subunit RPA190).