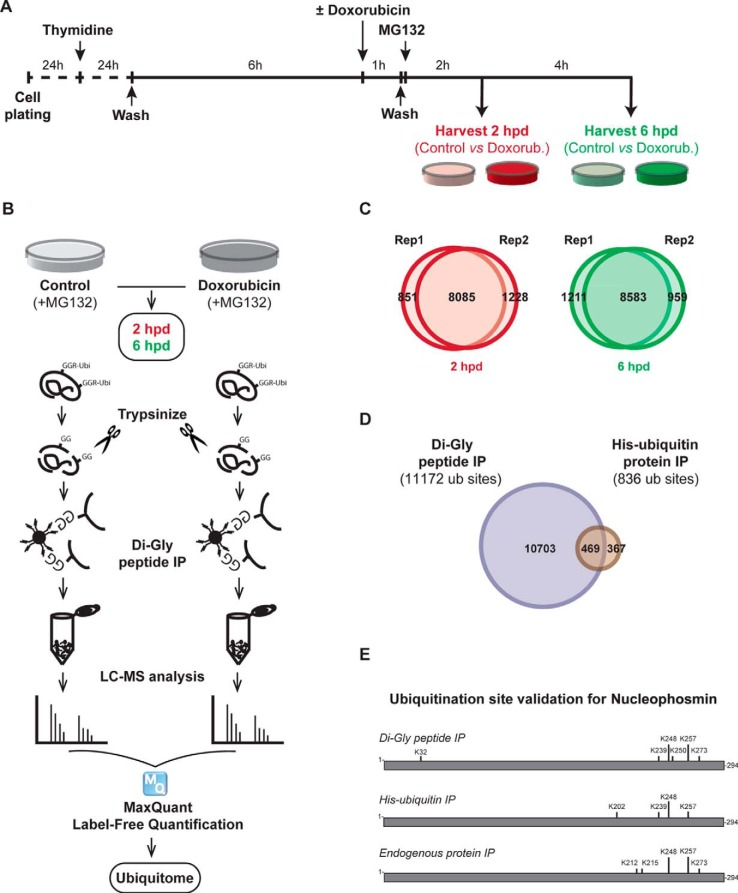
Fig. 1.
Experimental setting, proteomics analysis, and validation. (A) U2OS cells were synchronized in G2 using a thymidine block, followed by a 6-h release. Subsequently, DNA damage was induced by a 1-h doxorubicin pulse. MG132 was added after the pulse to inhibit proteasomal degradation. Cells were harvested 2 h and 6 h after DNA damage treatment for proteomics analysis. Two biological replicates were generated (B) proteomics platform. Following the harvest, cells were lysed and proteins were digested with trypsin. Dyglycil (di-Gly) peptides were enriched with ubiquitin remnant peptide IP. Peptides were analyzed with LC-MS, followed by MaxQuant label-free quantification. Three MS runs were performed and combined for each biological replicate. (C) Venn diagrams show the overlap between both biological replicates with respect to ubiquitin sites identified in the 2-h post-damage (left) and 6-h post-damage (right) time points. (D) Independent validation of identified ubiquitin sites. Venn diagram shows the number of ubiquitin sites identified following ubiquitin remnant peptide IP (blue) and His-ubiquitin protein IP (red). The overlap shows the number of ubiquitin sites that were identified after both enrichment methods. (E) Schematic representation of ubiquitin sites identified in NPM protein following di-Gly peptide IP, His-ubiquitin IP, and endogenous protein IP.