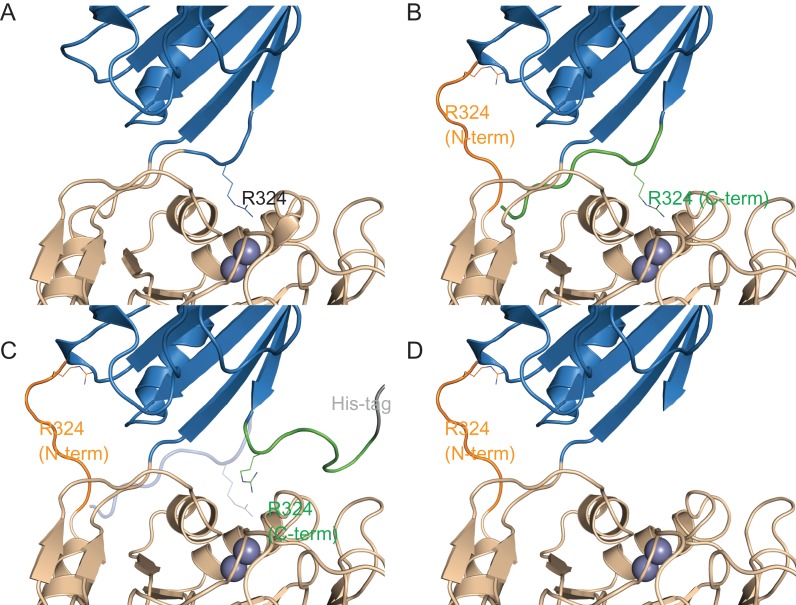
Fig. 8.
Structural basis for the difference in activity of the CPG2CP-323-330 series of circular permutants. The catalytic domain is shown in wheat, and the dimerization domain is shown in blue. The residue 323–330 sequence is shown in orange when attached to the N-terminus of the circular permutant, and in green when attached to the C-terminus. The position of catalytically important R324 is shown as sticks. The dinuclear zinc active site is shown as two spheres. The His-tag is shown in gray. Differences from the CPG2 crystal structure (PDB ID 1CG2) are for illustrative purposes only, and do not represent computationally modeled or experimentally determined structures. (A) Wild-type CPG2. (B) The CPG2CP-N323-C330-N-His circular permutant (His-tag is not shown). The C-terminal sequence is believed to occupy the ‘canonical’ position, allowing only a slight reduction of activity. The N-terminal sequence is shown adopting a random conformation, or in the case of CPG2CP-N330 (not shown), is deleted. (C) The CPG2CP-N323-C330-C-His circular permutant. The C-terminal His-tag disrupts the positioning of the C-terminal 323–330 sequence, preventing it from occupying the ‘canonical’ position (transparent blue cartoon). As the C-terminal sequence is not available to occupy the ‘canonical’ position and the N-terminal sequence is unable to compensate, the activity is reduced. (D) The CPG2CP-N323 circular permutant (His-tag is not shown). As the C-terminal sequence is not available to occupy the ‘canonical’ position and the N-terminal sequence is unable to compensate, the activity is greatly reduced.