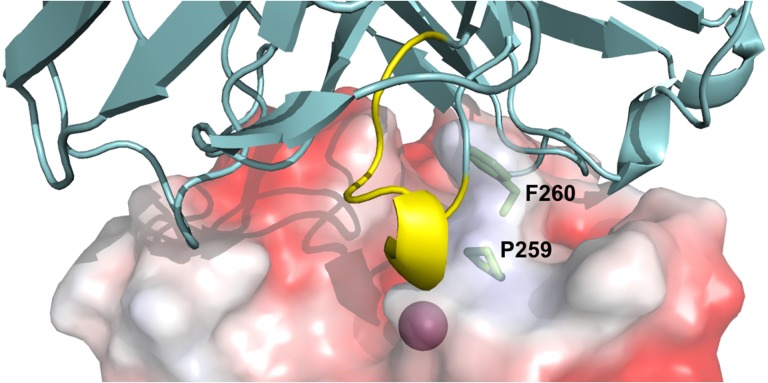
Fig. 5.
Structural prediction of Fab 1F8/cdMMP-14 complex. The active site of cdMMP-14 (electrostatic potential surface model, PDB ID 1bqq, Fernandez-Catalan et al., 1998) was shown with the catalytic Zn2+ (magenta) at the bottom of the reaction pocket. Fv 1F8 (cartoon, cyan) binds to vicinity of cdMMP-14 reaction cleft through direct interaction between the grafted Peptide G motif (yellow) and the active site. MMP-14 residues Pro259 and Phe260 (sticks, green) form the S1′ MMP-specific substrate binding pocket. Model of Fab 1F8 was generated using SAbPred (Dunbar et al., 2016). Fab 1F8 and cdMMP-14 were docked using ZDOCK (Pierce et al., 2014). Images were generated using PyMOL.