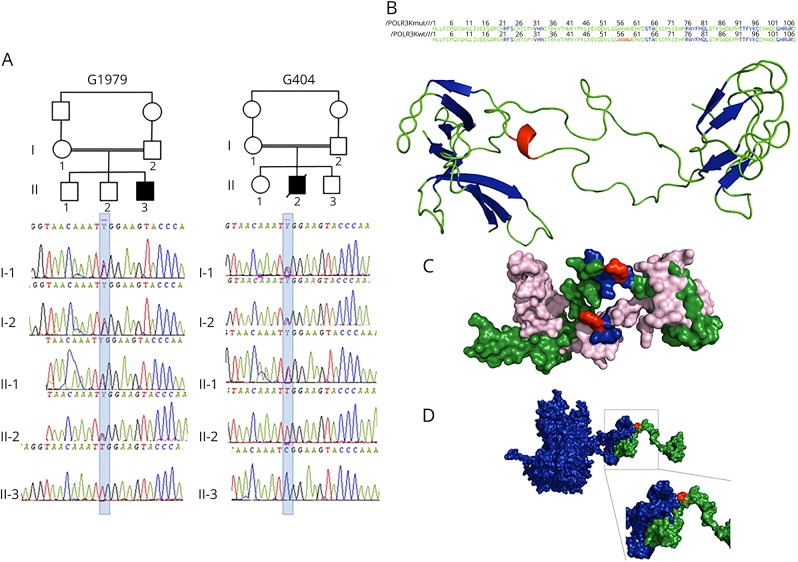
Figure 1. POLR3K mutated families, structural model of POLR3K and of POLR3K-POLR3B interactions.
(A) Pedigree and electropherograms of family G1979 (patient 1) and family G404 (patient 2). (B) Amino acid (AA) sequence and 2-dimensional structure of the mutated and wild-type POLR3K. The sheets, loops, and α-helix motifs are colored in blue, green, and red, respectively. The loop (AA 34–55), α-helix (AA 56–59), and loop (AA60–63) motifs of the wild-type protein are replaced by a unique loop (34–63) in the mutated protein. (C) Three-dimensional structure of the wild-type (in green) and mutated (in pink) POLR3K. The AAs at position 41 (arginine in the wild type and tryptophan in the mutant) are in red. The residues located within 4 Å around the Arg41, responsible for the stability of the protein (Asn40, Lys42, and Tyr43), are colored in blue. The Trp41 change induces a modification in the interactions of Tyr43 with Asn40 and Lys42 decreasing protein stability. (D) Three-dimensional structure of POLR3K (in green) and POLR3B (in blue) interactions. The mutated residue 41 colored in red is important in POLR3K-POLR3B interactions: the N-terminal part (1–41) of POLR3K interacting with the C-terminal part (1079–1133) of POLR3B.