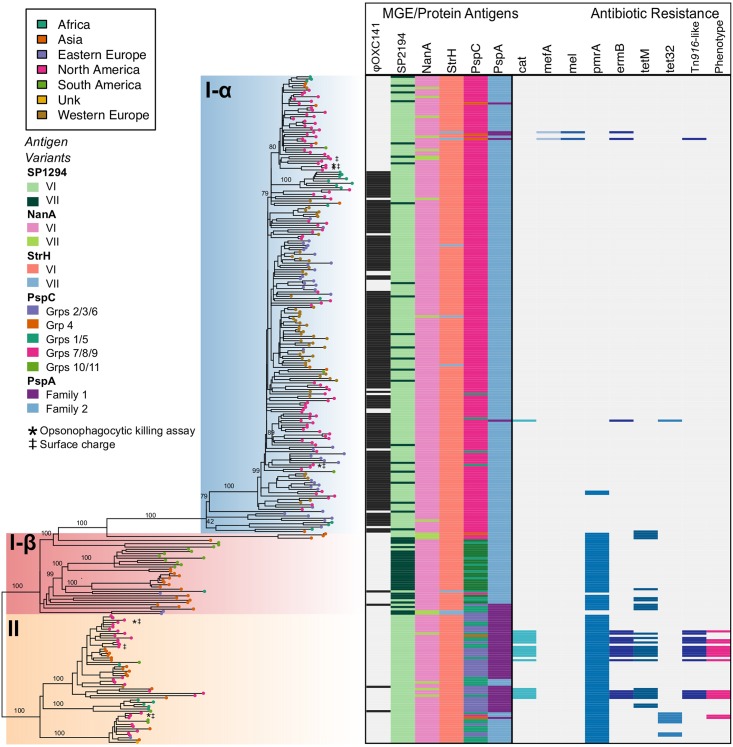
Fig 5. Phylogeny, polymorphic protein antigen variants, and antibiotic resistance.
Midpoint rooted maximum-likelihood phylogeny corresponding to Fig 2A with geographic region of isolation represented as colored tip shapes. Clades I-α, I-β, and II are shaded consistent with Fig 2A. Bootstrap values from 100 replicates are provided for major clades and sub-clades (see supplemental phylogeny for all values). Strains used in opsonophagocytic killing assay (asterisk) and surface charge experiments (double dagger) are indicated on the phylogeny. Corresponding protein antigen variants for SP1294, NanA, StrH, PspC, and PspA are illustrated on the left half of the heatmap. Eight other protein antigens are excluded due to a lack of variation in the sample. The presence and absence of AMR-associated genes are illustrated on the right half of the heatmap. The last column of the heatmap indicates genotypic antibiotic resistance that was confirmed by phenotypic testing (broth dilution or disk diffusion).