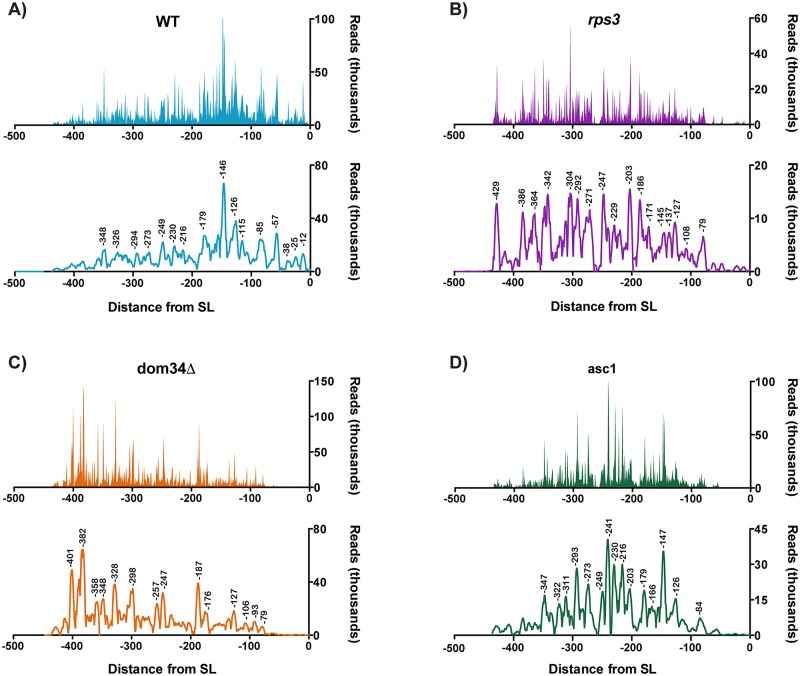
Fig 5. Large scale sequencing reveals changes in cleavage patterns in the presence of RPS3 mutations.
Plot of sequencing reads of 3’ RACE products from the indicated strains, each expressing PGK1-SL. All strains are in a ski2Δ background. Each point represents a single read, mapped relative to the stall site and the bottom plot in each panel denotes smoothed data, produced using a 5-point quadratic polynomial. Peaks with values at the 75th quartile or above are labelled as position relative to the stall sequence. (A-B) Compared to wild type (A), RPS3 mutations (B) result in highly heterogeneous cleavage without a predominant peak. (C-D) Deletion of DOM34 (C) or mutations in ASC1 (D) each produce greater heterogeneity of cleavage products with distinct patterns compared to each other and to wild type. Data in panel A is adapted from Simms et al [62].