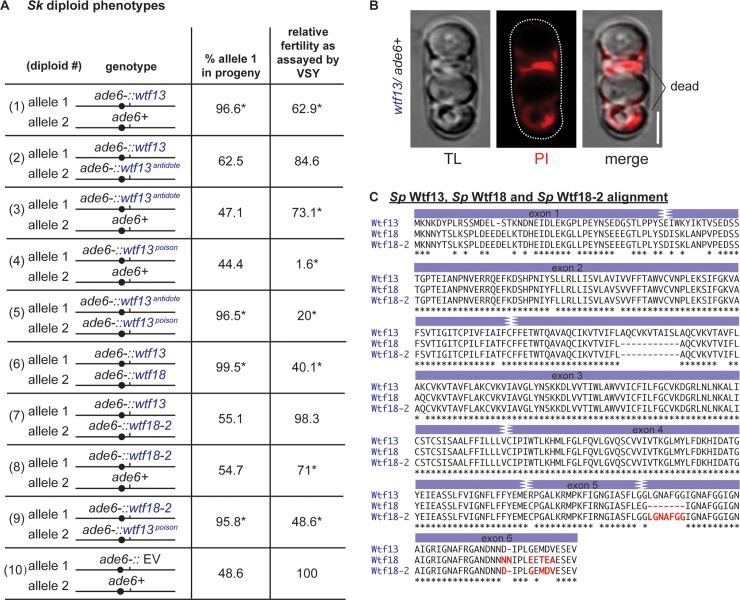
Fig 2. Sp wtf13 is a meiotic driver that is suppressed by Sp wtf18-2.
(A) Allele transmission and fertility of Sk diploids with the indicated constructs integrated at the ade6 marker locus. Alleles were followed using drug resistance markers (kanMX4 or hphMX6). Diploids 1–9 were compared to a wild-type empty-vector (EV) control (diploid 10) to detect biased allele transmission and differences in fertility as measured by viable spore yield. * indicates p-value of < 0.05 (G-test [allele transmission] and Wilcoxon test [fertility]). Fertility was normalized to the empty-vector control and reported as percent. More than 200 viable haploid spores were genotyped for each cross. Spores that inherited both markers and were thus geneticinR Ade+, hygromycinR Ade+ or hygromycinR geneticinR were excluded from the analyses. Raw data can be found in S1 and S2 Tables. (B) Transmitted light (TL) and fluorescent images from a tetrad produced by a wtf13/ade6+ heterozygous diploid (diploid 1) stained with propidium iodide (PI). Dead cells that lose membrane integrity stain with PI, but viable cells exclude the dye. 86% of stained tetrads exhibited this pattern (n = 58, S4 Fig). The scale bar represents 3μm. (C) Amino acid sequence alignment of Sp Wtf13 (long isoform), Sp Wtf18 and Sp Wtf18-2. In red are the amino acid differences between Wtf18 and Wtf18-2. Those red residues are all identical between Sp Wtf13 and Sp Wtf18-2.