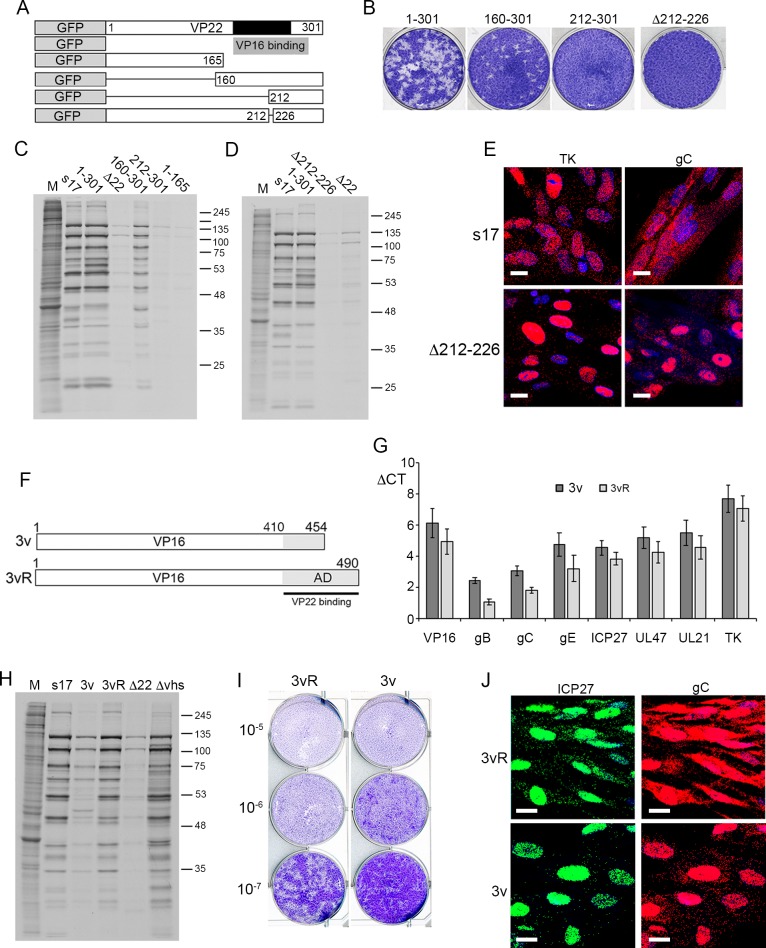
Fig 10. Cytoplasmic localisation of late transcripts is enhanced by VP22 binding to VP16.
(A) Line drawing of VP22 variants expressed as GFP fusion proteins in virus infection. Black box indicated the conserved domain of VP22, grey box indicates the region required for VP16 binding. (B) Plaque formation of viruses shown in A on HFFF cells. (C) & (D) HFFF cells were infected with Wt (s17), Δ22 or the viruses shown in A at a multiplicity of 2, were metabolically labelled with [35S]-methionine 15 hours after infection. Cells were lysed and analysed by SDS-PAGE and autoradiography. (E) HFFF cells infected with Wt (s17) or Δ212–226 at a multiplicity of 2 were fixed at 16 hours and subjected to mRNA FISH with probes specific for the IE transcript ICP27 or the late transcript gC (both in red). Nuclei were counterstained with DAPI (blue). Scale bar = 20 μm. (F) Line drawing of the variant of VP16 (Δ454–490) expressed in the 3v virus based on the KOS strain, together with its rescue virus 3vR. The grey box indicates the C-terminal activation domain (AD) of VP16, the black line indicates the region of VP16 required to bind VP22. (G) HFFF cells infected with the viruses shown in F at a multiplicity of 2 were harvested for total RNA at 16 hours and analysed by qRT-PCR for the indicated transcripts. Results are represented as ΔCT values with the mean ± standard error for n = 3 shown. (H) HFFF cells infected with Wt (s17), Δ22, Δvhs or the viruses shown in F at a multiplicity of 2, were metabolically labelled with [35S]-methionine 15 hours after infection. Cells were lysed and analysed by SDS-PAGE and autoradiography. (I) The 3v and 3vR viruses were titrated onto HFFF cells and plaques fixed and stained with crystal violet 5 days later. (J) HFFF cells infected with 3v or 3vR viruses at a multiplicity of 2 were fixed at 16 hours and subjected to multiplex mRNA FISH with probes specific for the IE transcript ICP27 (green) and the late transcript gC (red). Nuclei were counterstained with DAPI (blue). Scale bar = 20 μm.