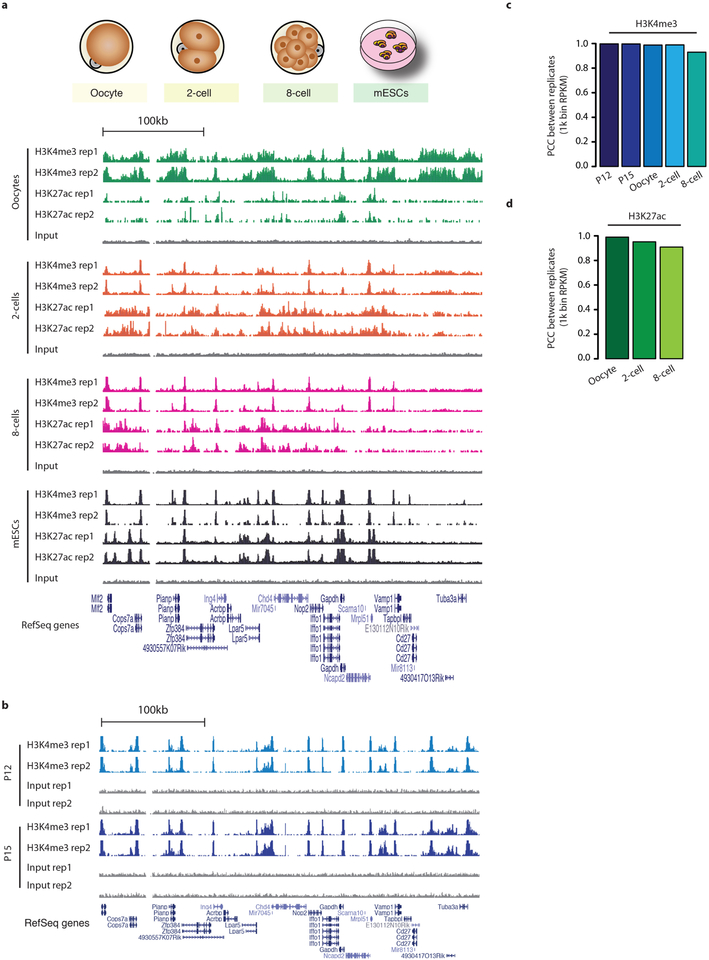
Extended Data Figure 2 |. Reproducibility of μChIP-seq experiments.
a, Genome browser snapshots of H3K4me3 and H3K27ac μChIP-seq results in MII oocytes, 2- and 8-cell embryos and ES cells. For each histone-modification mark, we generated two biological replicates. b, Genome browser snapshots of H3K4me3 μChIP-seq results from P12 and P15 oocytes. We generated two biological replicates. c, d, Bar plots show the Pearson correlation coefficient between two biological replicates for H3K4me3 (c) and H3K27ac (d). Pearson correlation coefficients were calculated using RPKM values for 1-kb-binned regions of the whole genome.