Figure 4. Tak1 is necessary for baseline vesicle fusion and PHP under physiological conditions.
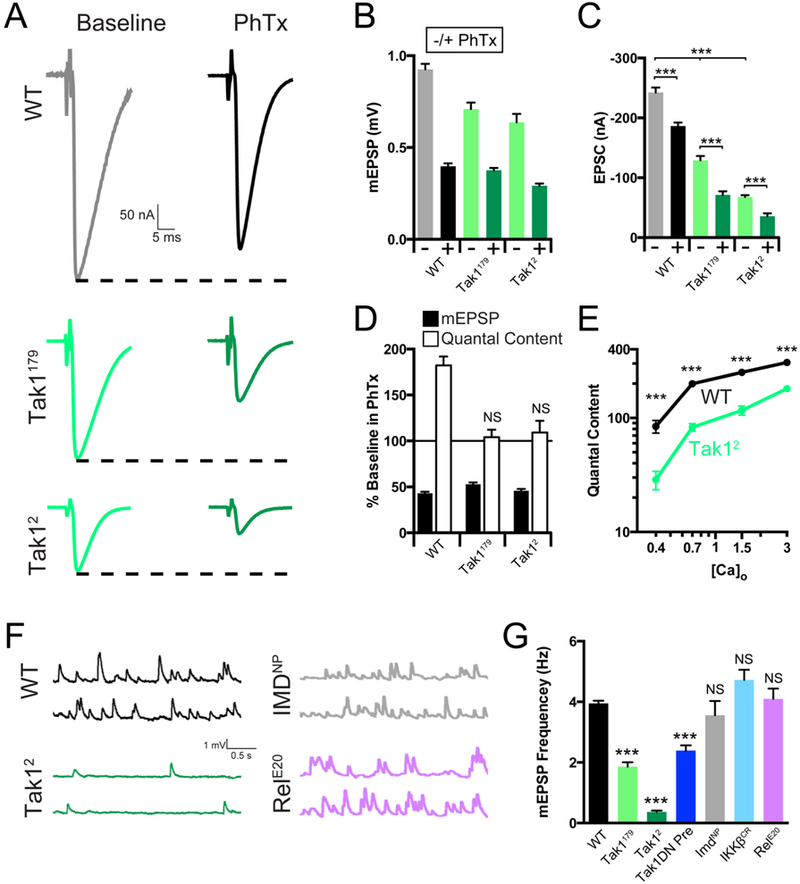
(A) Representative traces for EPSC (scale, 50 nA, 5 ms) at baseline, and in the presence of PhTx, for the indicated genotypes. (B) Average mEPSP amplitude for each genotype in the absence (light bars) or presence (dark bars) of PhTx. (C) Average EPSC amplitude in the absence (light bars) or presence (dark bars) of PhTx. (D) mEPSP amplitudes (filled bars) and quantal content (open bars) for each genotype in the presence of PhTx, normalized to baseline values in the absence of PhTx. (E) Calcium cooperativity curves for the indicated genotypes. Neurotransmitter release was measured at 0.4, 0.7, 1.5, and 3mM extracellular calcium concentration. Quantal content was calculated by dividing EPSC amplitudes by mEPSC amplitudes. (F) Representative traces for mEPSP (scale, 1 mV, 0.5 s) for the indicated genotypes. (G) Average mEPSP frequency for the indicated genotypes. Data are presented as average (+/− SEM) and statistical significance determined by Student’s t-test (unpaired, two-tailed) (C) or by one-way ANOVA with Tukey’s multiple comparisons test (G).
