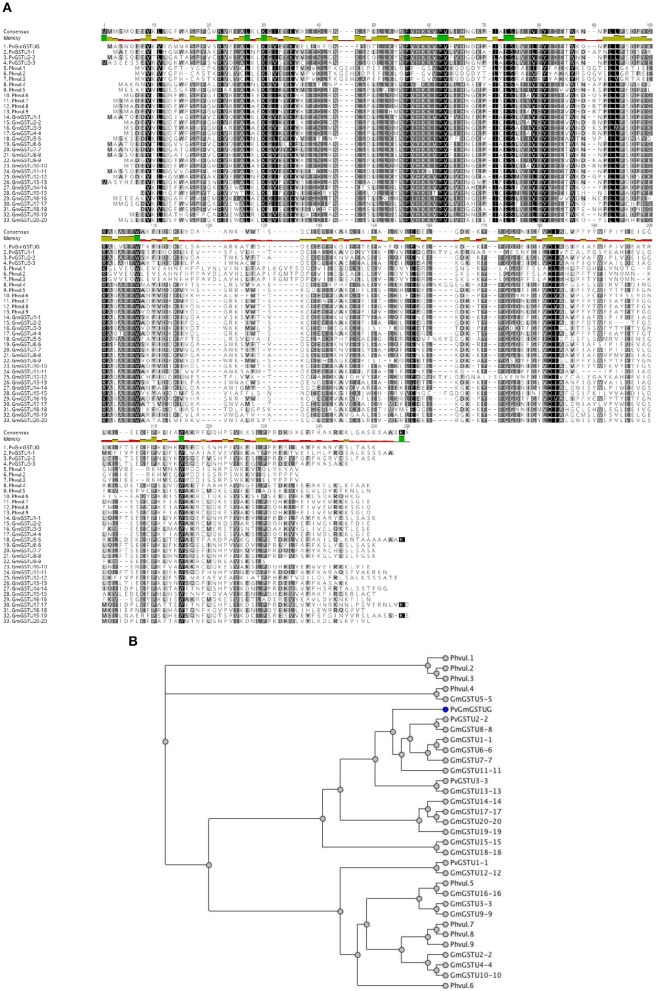
Figure 5.
Sequence and phylogenetic analysis (A). Amino acid sequence alignments of PvGmGSTUG with the tau class GSTs from Glycine max and Phaseolus vulgaris
(B). Phylogenetic analysis of GmPvGSTUG with the tau class GSTs from Glycine max and Phaseolus vulgaris. Phylogenetic tree was constructed by the neighbor joining method using Geneious v9.1.2 software (Kearse et al., 2012) after alignment of the protein sequences using the Clustal Omega sequence alignment program (Sievers et al., 2011). The figures were created using Geneious v9.1.2 software (Kearse et al., 2012). Conserved areas are shown shaded:  100% identity,
100% identity,  80–100% identity,
80–100% identity,  60–80% identity, < 60% identity. The accession numbers and gene codes of the GST sequences that were used were: PvGSTU1-1 (AEX38000.1); PvGSTU2-2 (AEX38001.1); PvGSTU3-3 (NP_171792); Phvul.1 (006G023500.1|PACid:27165305); Phvul.2 (008G195500.1|PACid:27155547); Phvul.3 (008G195600.1|PACid:27155113); Phvul.4 (002G080200.1|PACid:27169916); Phvul.5 (005G053300.1 PACid:27149482); Phvul.6 (005G053200.1|PACid:27149239); Phvul.7 (005G054000.1|PACid:27150418); Phvul.8 (code 005G054100.1|PACid:27148744); and Phvul.9 (005G054200.1|PACid:27149131). The accession numbers of Glycine max GST sequences that were used were: GmGSTU1-1, AAA33973; GmGSTU2-2, CAA71784; GmGSTU3-3, CAA48717; GmGSTU4-4, AAC18566; GmGSTU5-5, AAG34795; GmGSTU6-6, AAG34796; GmGSTU7-7, AAG34797; GmGSTU8-8, AAG34798; GmGSTU9-9, AAG34799; GmGSTU10-10, AAG34800; GmGSTU11-11, AAG34801; GmGSTU12-12, AAG34802; GmGSTU13-13, AAG34803; GmGSTU14-14, AAG34804; GmGSTU15-15, AAG34805; GmGSTU16-16, AAG34806; GmGSTU17-17, AAG34807; GmGSTU18-18, AAG34808; GmGSTU19-19, AAG34809; and GmGSTU20-20, AAG34810.
60–80% identity, < 60% identity. The accession numbers and gene codes of the GST sequences that were used were: PvGSTU1-1 (AEX38000.1); PvGSTU2-2 (AEX38001.1); PvGSTU3-3 (NP_171792); Phvul.1 (006G023500.1|PACid:27165305); Phvul.2 (008G195500.1|PACid:27155547); Phvul.3 (008G195600.1|PACid:27155113); Phvul.4 (002G080200.1|PACid:27169916); Phvul.5 (005G053300.1 PACid:27149482); Phvul.6 (005G053200.1|PACid:27149239); Phvul.7 (005G054000.1|PACid:27150418); Phvul.8 (code 005G054100.1|PACid:27148744); and Phvul.9 (005G054200.1|PACid:27149131). The accession numbers of Glycine max GST sequences that were used were: GmGSTU1-1, AAA33973; GmGSTU2-2, CAA71784; GmGSTU3-3, CAA48717; GmGSTU4-4, AAC18566; GmGSTU5-5, AAG34795; GmGSTU6-6, AAG34796; GmGSTU7-7, AAG34797; GmGSTU8-8, AAG34798; GmGSTU9-9, AAG34799; GmGSTU10-10, AAG34800; GmGSTU11-11, AAG34801; GmGSTU12-12, AAG34802; GmGSTU13-13, AAG34803; GmGSTU14-14, AAG34804; GmGSTU15-15, AAG34805; GmGSTU16-16, AAG34806; GmGSTU17-17, AAG34807; GmGSTU18-18, AAG34808; GmGSTU19-19, AAG34809; and GmGSTU20-20, AAG34810.