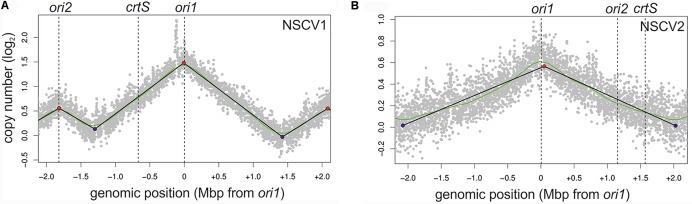
FIGURE 3.
Marker frequency analysis (MFA) of V. cholerae NSCV1 (A) and NSCV2 (B) to assess origin activities. Profiles of genome-wide copy numbers based on Illumina sequencing and read mapping. Gray dots represent log numbers of normalized reads as mean values for 1 kbp windows relative to the stationary phase sample. The genome position is shown as the distance from ori1. Vertical dotted black lines mark the locations of replication origins and the crtS site. The solid black lines represent the fitting of regression lines and the green line corresponds to the Loess regression (F = 0.05). Maxima are highlighted by red and minima as blue dots. Plots of biological replicates are shown in Supplementary Figure S1.