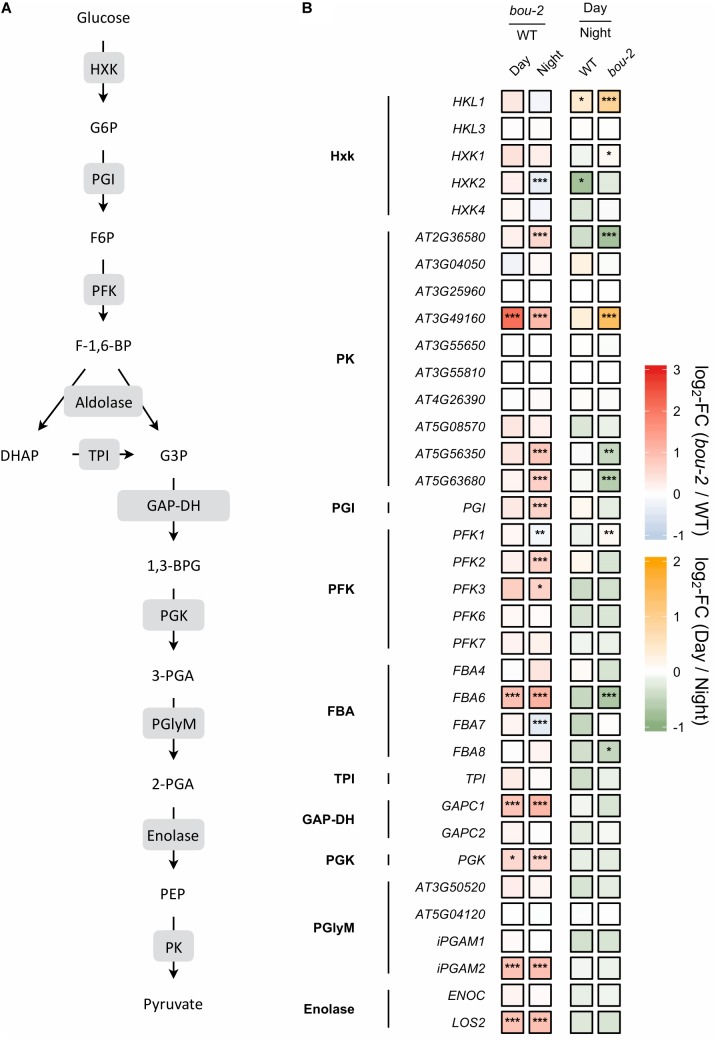
FIGURE 5.
(A) Schematic presentation of glycolysis. (B) Transcript levels of genes encoding for enzymes highlighted in gray boxes in panel A. Left two tiles of each gene represent the log2-fold change (FC) between the bou-2 mutant and WT plants under both day (D) and night (N) with false color code ranging from red (higher in bou-2) to blue (higher in WT). Right two tiles represent the log2-fold change (FC) between day and night within WT plants and the bou-2 mutant with false color code ranging from green (higher at night) to yellow (higher at day). Asterisks indicate differential gene expression as determined by Sleuth (∗FDR < 0.05, ∗∗FDR < 0.01, and ∗∗∗FDR < 0.001). HXK, hexokinase; PGI, phosphoglucose isomerase; PFK, phosphofructokinase; FBA, fructose-bisphosphate aldolase; TPI, triosephosphate isomerase; GAP-DH, glyceraldehyde-3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; PGlyM, phosphoglycerate/bisphosphoglycerate mutase; PK, Pyruvate kinase.