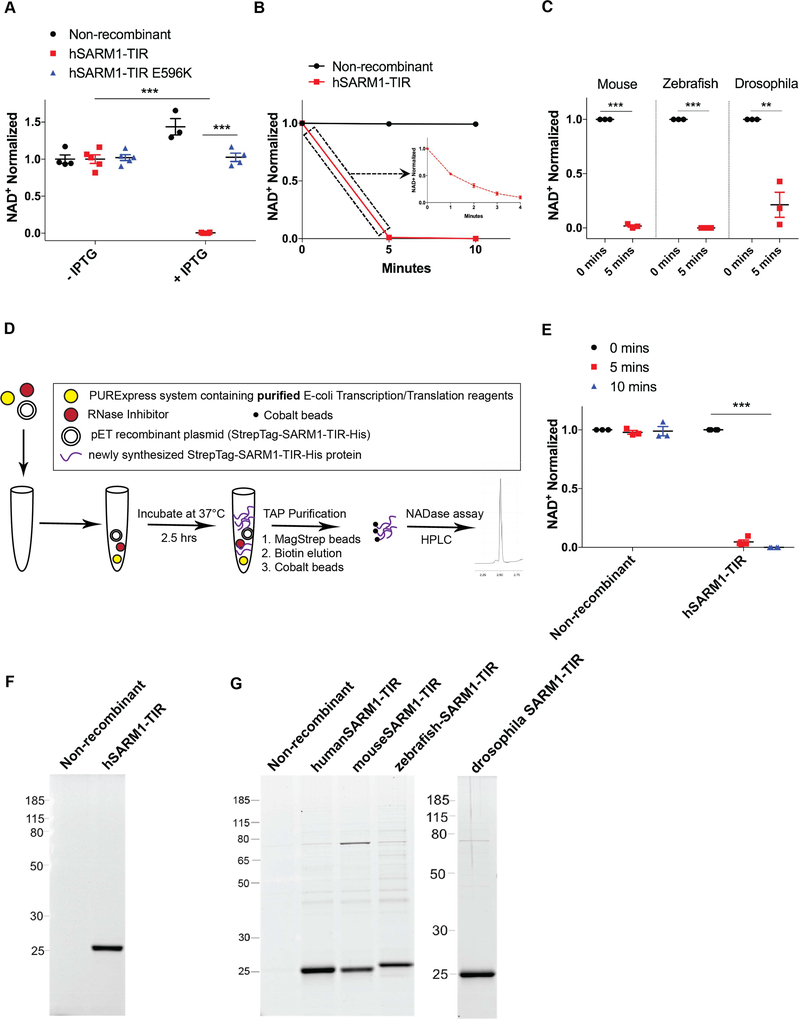
Figure 2: NAD+ cleavage enzymatic activity is intrinsic to SARM1-TIR.
A) Endogenous NAD+ levels in bacteria after IPTG induction of human SARM1-TIR. B) In vitro NAD+ cleavage reaction by human SARM1-TIR protein expressed and purified from bacteria. C) Bacterially expressed mouse, zebrafish, and drosophila SARM1-TIR proteins cleave NAD+ in NADase assay. D) Schematic of cell-free protein expression system. E) Human SARM1-TIR purified from cell-free protein expression system cleaves NAD+ in NADase assay. F) SYPRO Ruby gel of SARM1-TIR laden beads purified from cell-free transcription/translation system. G) SYPRO Ruby gel of SARM1-TIR laden beads purified from bacteria. These cell-free and bacterially expressed proteins lack the Venus fluorescent tag and thus run at a different size than the proteins expressed in NRK1-HEK293T cells (compare to Figure 1). Data was generated from at least three independent reaction experiments using purified protein from at least three independent bacteria clones. Data are presented as mean ± SEM; Error bars: SEM; **P<0.01, ***P<0.001 unpaired two tailed Student’s t-test and one-way ANOVA for multiple comparisons. See also Figure S2.