Figure 1.
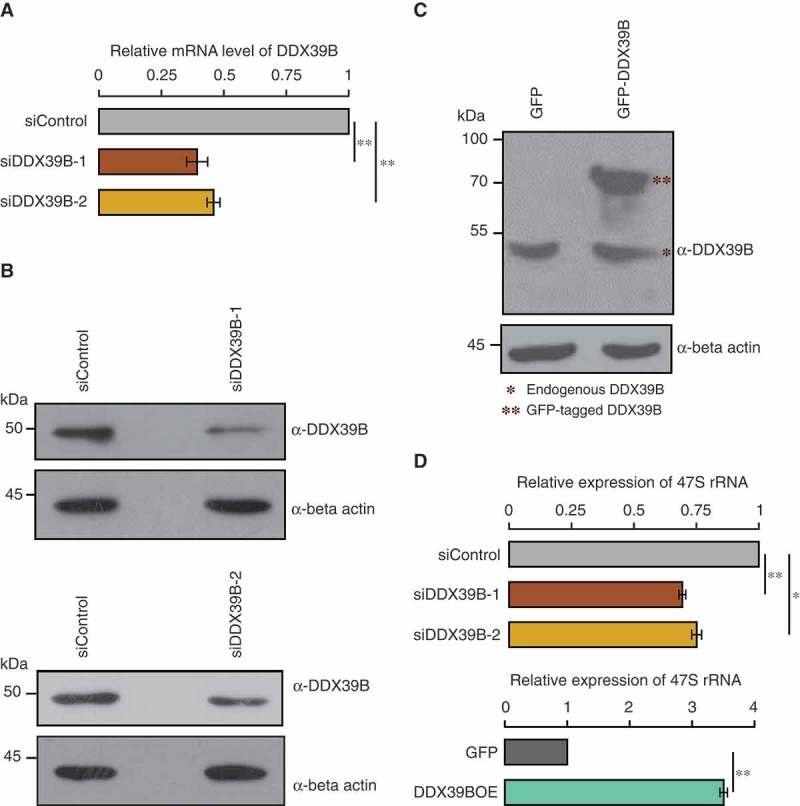
DDX39B regulates the levels of pre-ribosomal RNA.
(A) Efficiency of siRNA mediated knockdown of DDX39B by qRT-PCR. HEK293 cells were treated with control siRNA or DDX39B siRNA-1 (siDDX39B-1) or DDX39B siRNA-2 (siDDX39B-2) and transcript levels of DDX39B were quantified using qRT-PCR. DDX39B transcripts levels were normalized to GAPDH expression and are presented relative to the control sample. Data are represented as mean of three independent experiments, with error bars representing standard deviation. Statistical significance was assessed by two tailed t-Test: paired two samples for means. ** represents P-value <0.01. (B) DDX39B protein levels in HEK293 cells treated with control siRNA or siDDX39B-1 (Upper panel) or siDDX39B-2 (Lower panel) were analysed using western blotting. The immunoblotting was performed with whole cell extracts using DDX39B antibody or beta actin antibody. (C) Comparison of over-expressed DDX39B with endogenous DDX39B. HEK293 cells were transfected with pEGFP-C1 vector or pEGFP-DDX39B construct and the DDX39B levels were analysed using western blotting. The western blotting was performed with whole cell extracts using DDX39B antibody or beta actin antibody. (D) DDX39B maintains the steady state levels of pre-ribosomal RNA. DDX39B levels were perturbed in HEK293 cells by transfecting with siDDX39B-1 or siDDX39B-2 or pEGFP-DDX39B and pre-ribosomal 47S rRNA levels were quantified by qRT-PCR. The 47S rRNA levels were normalized to GAPDH expression and are presented relative to the control sample. Data are represented as mean of three independent experiments, with error bars representing standard deviations. Statistical significance was assessed by two tailed t-Test: paired two samples for means. * represents P-value <0.05 and ** P-value <0.01.
