Figure 2.
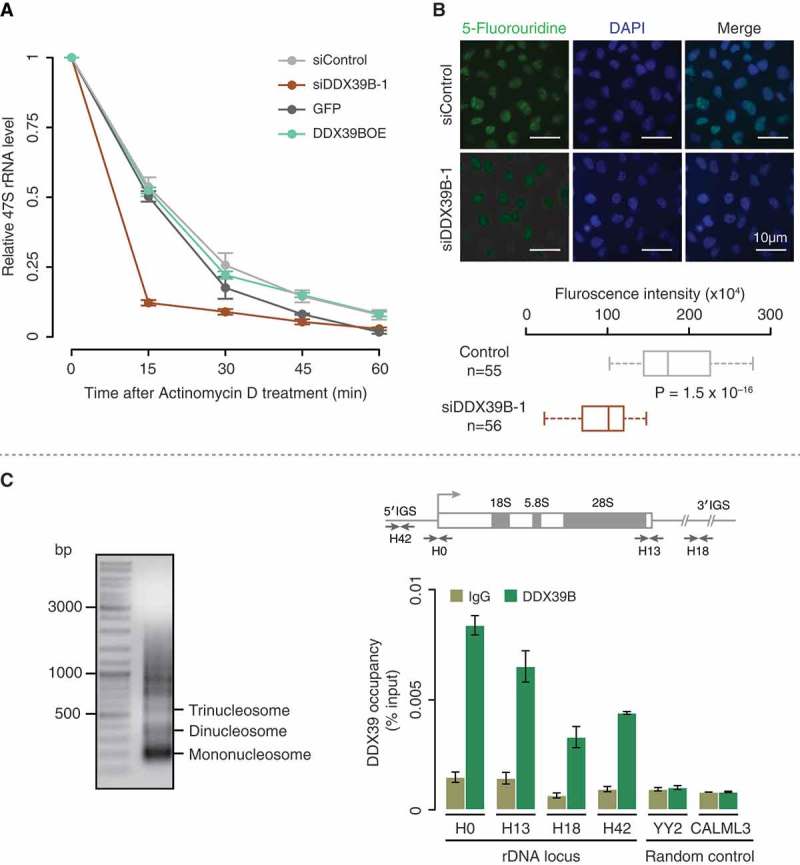
DDX39B regulates the stability and transcription of pre-ribosomal RNA.
(A) DDX39B perturbation affects the stability of pre-ribosomal RNA. HEK293 cells were transfected with control or siDDX39B-1 or pEGFP-C1 vector or pEGFP-DDX39B. After 48 hours of transfection, the cells were harvested at the indicated time points after the addition of actinomycin D. The levels of 47S rRNA were then quantified by qRT-PCR. The 47S rRNA levels were normalized to GAPDH expression and are presented relative to the 0 min time point sample. Data are represented as mean of three independent experiments. The error bars represent standard deviations. (B) DDX39B is required for efficient synthesis of pre-ribosomal rRNA. The control or DDX39B knock down HeLa cells were pulse labelled with 5-fluorouridine (5-FUrd) and immunostained with BrdU antibody (left panel). DNA was stained with DAPI (middle panel). The right panel shows the merge of fluorescent images. The graph at the bottom of the panel represents the quantification of fluorescence signals in 55 cells of each group. The quantification of fluorescence signals was done by using ImageJ software. Statistical significance was assessed using nonparametric Wilcoxon rank-sum test. P-Values are depicted. (C) DDX39B is recruited to the promoter and regulatory regions of rDNA. The chromatin was prepared from HEK293 cells and fragmented (upper panel gel image). Immunoprecipitation was performed using rabbit IgG antibody or DDX39B antibody. The immunoprecipitated DNA was investigated for the enrichment of promoter and regulatory regions of rDNA and two random control regions (YY2 and CALML3) by using qRT-PCR. The primer binding locations in the rDNA locus are indicated in the schema (above the graph). Data are shown relative to input and the values represent the mean of three independent experiments, with the error bars depicting standard deviations.
