Figure 5.
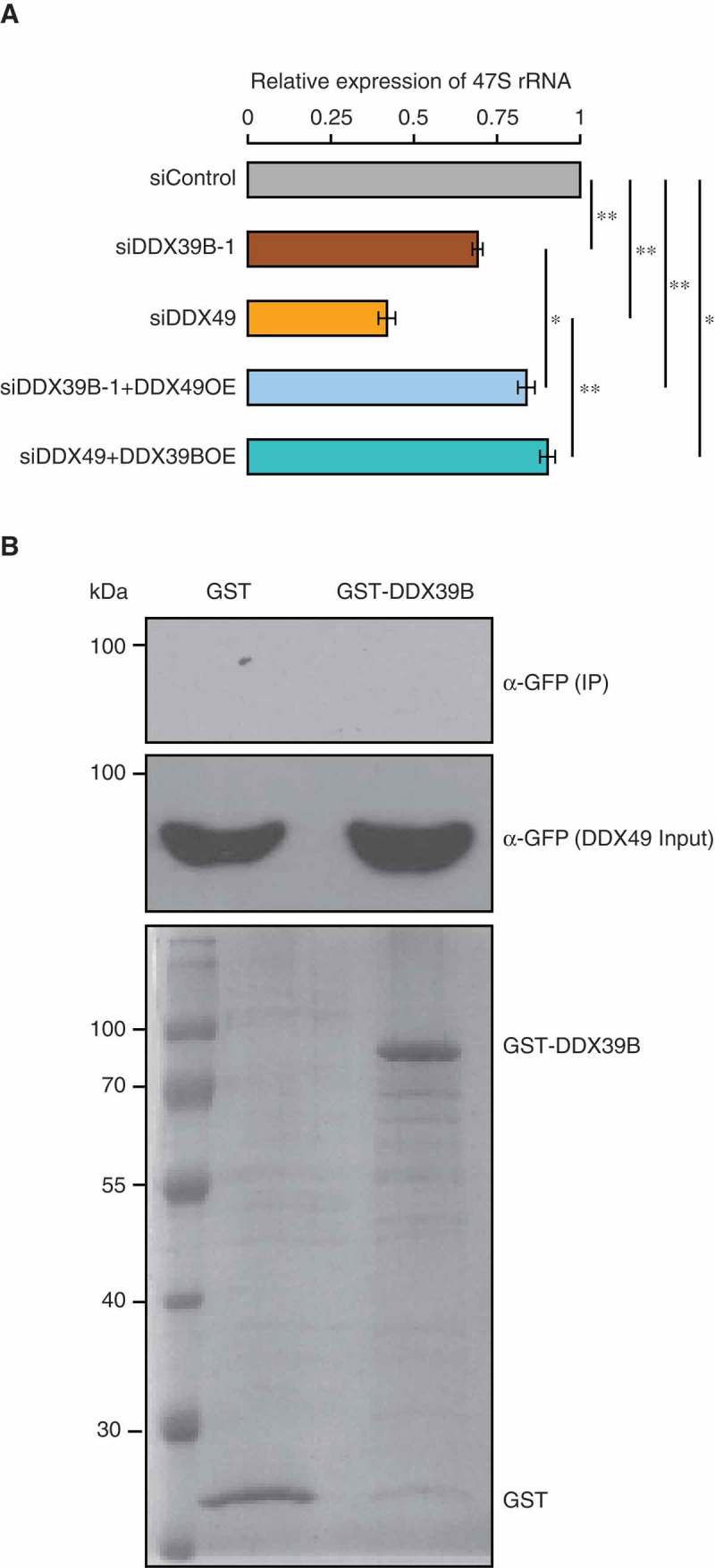
DDX39B partly compensates the function of DDX49 and vice versa.
(A) Functional compensation of DDX39B by DDX49 and vice versa. HEK293 cells were transfected with control siRNA or siDDX39B-1 or DDX49 siRNA or combination DDX49 siRNA and pEGFP-DDX39B or combination siDDX39B-1 and pEGFP-DDX49 and the 47S rRNA levels were quantified using qRT-PCR. The 47S rRNA levels were normalized to GAPDH expression and are presented relative to the control sample. Data are represented as mean of three independent experiments, with error bars representing standard deviations. The 47S rRNA levels were normalized to GAPDH expression and are presented relative to the control sample. Data are represented as mean of three independent experiments, with error bars representing standard deviations. Statistical significance was assessed by two tailed t-Test: paired two samples for means. * represents P-value <0.05 and ** P-value <0.01.(B) DDX39B does not interact with DDX49. The possibility of DDX39B-DDX49 interaction was investigated by GST pulldown assay. The GST-tagged DDX39B or the GST protein were coupled to the glutathione sepharose and the beads were incubated with the cell lysates prepared from the cells, which were overexpressing of GFP-tagged DDX49 protein. The beads were washed with wash buffer and the bound fractions were probed with anti-GFP antibody (Upper blot). About 1% of the whole cell lysates used in the pull-down assay, were probed with anti-GFP antibody (Middle blot) and 5% of the GST or GST-tagged DDX39B proteins, which were used in pull down assay were separated in SDS-PAGE and stained with coomassie blue dye (Lower blot).
