Figure 1.
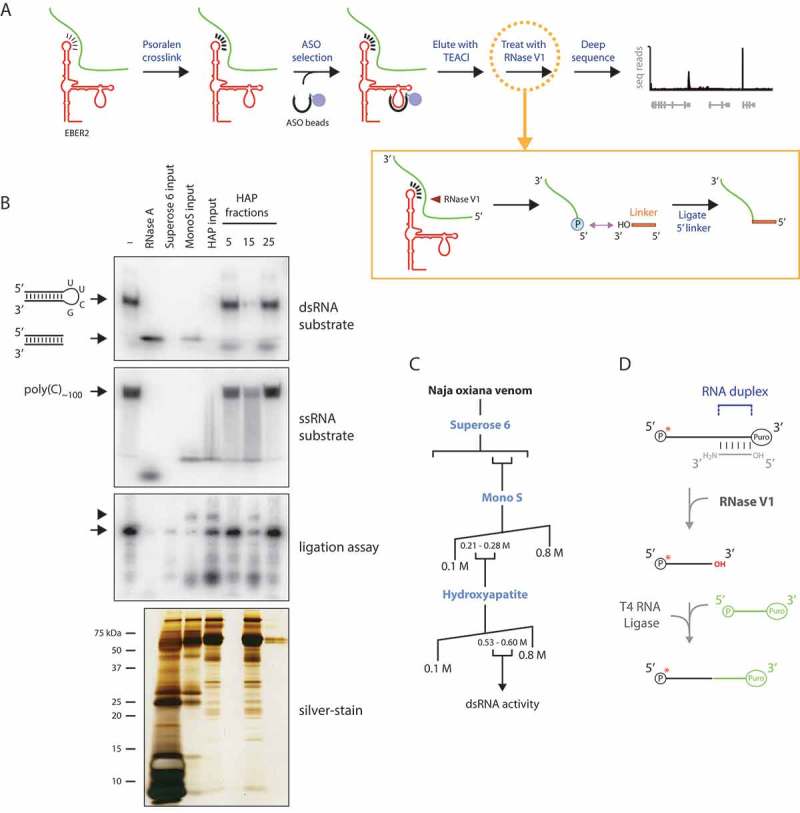
Identifying EBER2-interacting RNAs by combining psoralen crosslinking, ASO-mediated selection, and RNase V1 treatment. (A) The psoralen derivative AMT is used to crosslink RNA duplexes in intact cells to preserve in vivo RNA-RNA interactions. An EBER2-targeting ASO is then used to select EBER2 together with crosslinked interacting RNAs. These duplexes are eluted from the ASO beads using TEACl-containing buffer and are subjected to RNase V1 digestion. Following cleavage of double-stranded regions, a linker is ligated to the newly-generated 5′ phosphate group at the cut site using T4 RNA ligase (inset). Only one possible cleavage event is depicted for simplicity. After deep sequencing, not only can the interacting RNAs be identified, but also the site of RNA-RNA interactions can be deduced, which are specified by the junction of the linker and interacting RNA. (B) Cobra venom fractions were examined for activity towards doubled-stranded and single-stranded substrates. The double-stranded substrate consists of a shRNA with a pyrimidine-rich loop, which can be digested by single-strand specific RNases, such as RNase A. The trimmed RNA duplex with no loop region migrates faster in a native polyacrylamide gel. Digestion within the stem region by a double-strand specific RNase results in the disappearance of radioactive signal, as observed after digestion with all input material as well as hydroxyapatite (HAP) fraction 15; note that the weak activity of the MonoS input sample is due to the great dilution of protein concentration following size exclusion chromatography. Indicated fractions were also used in a ligation assay (outlined in D) to verify the compatibility of RNase V1 digest with T4 RNA ligase reaction. A silver-stained gel of the purified fractions is shown in the bottom panel, revealing the partial purification only of RNase V1; many other proteins are present in our sample preparation, which, importantly, do not interfere with RNase V1 activity. (C) Purification scheme of RNase V1 from Naja oxiana venom. (D) Outline of ligation reaction after RNase V1 digest. An oligonucleotide blocked at the 3′ end with puromycin was 5′ end-labeled (arrow in B, third panel from top) and annealed to a partially complementary oligonucleotide with a 3′ amino modifier. A free 3′ OH group is created only after RNase V1 digest, to which a 5′ phosphorylated linker blocked at the 3′ end with puromycin can be ligated using T4 RNA ligase. This ligation product is the only one that can be visualized by autoradiography as shown in B (arrowhead, third panel from top).
