Figure 3.
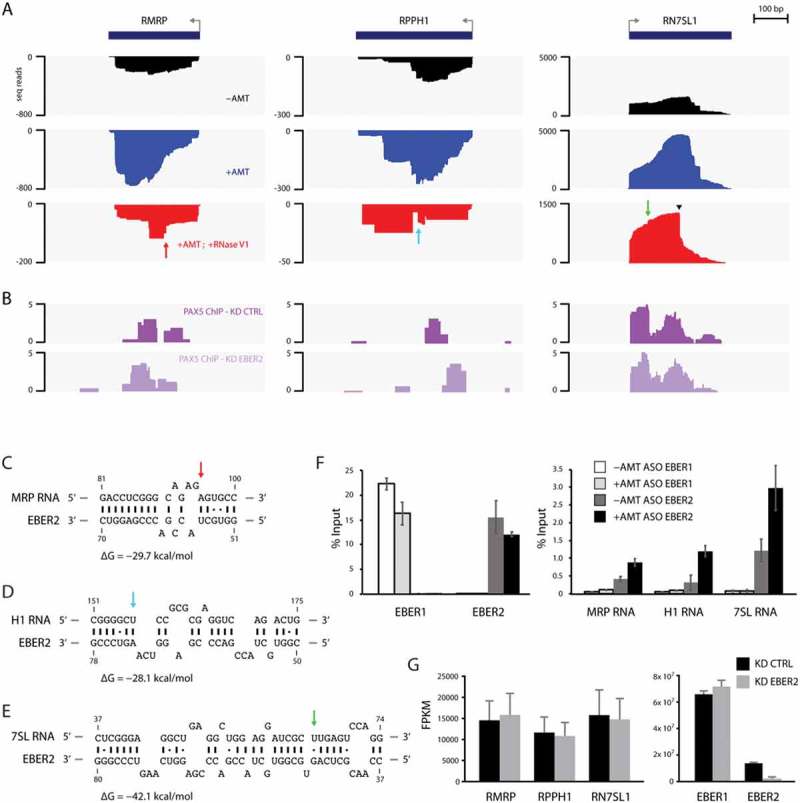
Candidate host EBER2-interacting RNAs. (A) Tracks for the gene loci RMRP, RPPH1 and RN7SL1 are shown after ASO-selection. All three functional noncoding RNAs exhibit increased coverage after AMT-crosslinking compared to ASO-selection of untreated total RNA. They also exhibit a distinct alignment profile when comparing the AMT-crosslinked sample to the one including RNase V1-digest. The latter shows abrupt edges in their profiles, indicative of RNA duplex formation. (B) PAX5 ChIP tracks of control (PAX5 ChIP–KD CTRL) and EBER2 knockdown (PAX5 ChIP–KD EBER2) BJAB-B1 cells. (C-E) Predicted RNA duplexes generated by the program RNAhybrid between the top stem-loop region of EBER2 and MRP (C), H1 (D), and 7SL RNAs (E). The calculated free energies of these duplexes are shown. Arrows indicate the abrupt edges shown in the sequencing tracks. (F) Enrichment of candidate host RNAs after AMT-crosslinking and EBER2 selection determined by quantitative RT-PCR. ASO targeting EBER1 was used as a negative control. While both EBER1 and EBER2 are precipitated at comparable levels, co-selection for MRP, H1, and 7SL RNA is only observed for ASO EBER2 and enhanced following AMT-crosslinking. Error bars indicate standard deviation of three independent experiments. (G) EBER2 depletion does not affect steady-state RNA levels of MRP, H1, and 7SL RNAs. FPKM values of the functional noncoding RNAs in control (KD CTRL) and EBER2 knockdown (KD EBER2) BJAB-B1 cells as determined by Cufflinks. Error bars indicate confidence intervals calculated by Cufflinks.
