Figure 1.
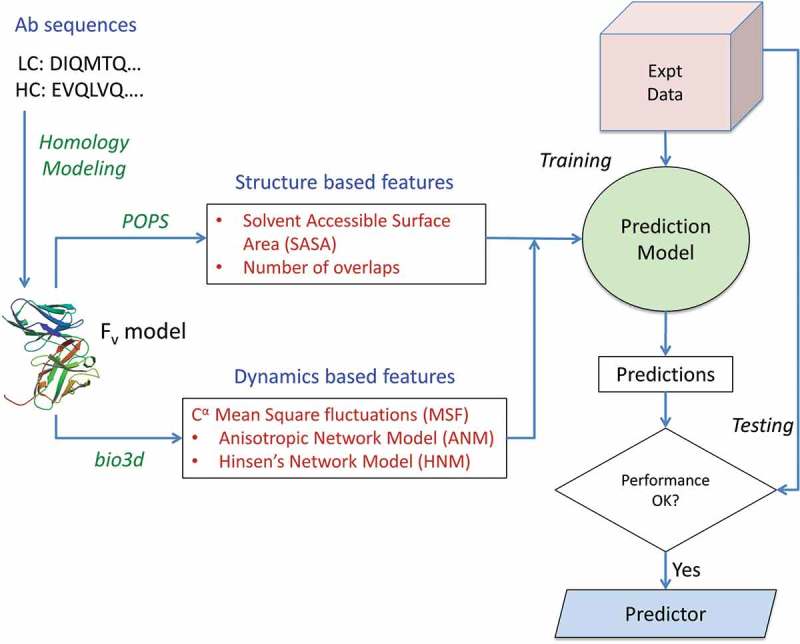
Schematic workflow of the methodology. Antibody sequences are obtained from an in-house database and the Fv regions for each structure modeled using MOE protocols. Features are extracted from the sequence, structure and dynamics of the mAb Fv regions and used to implement a random forest-based predictor in R. The performance of the model is assessed using the standard metrics of correlation and root mean square error for regressor model and accuracy, precision, sensitivity and specificity for the implicit classifier model.
