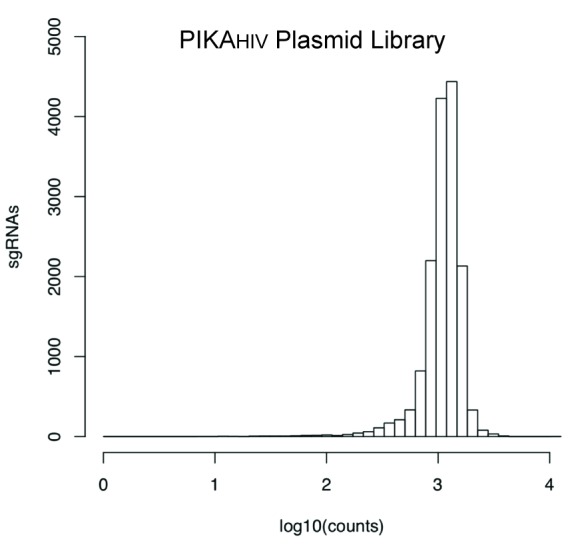
Figure 1. The HIV-CRISPR screen identifies gene knockouts that increase and decrease HIV infection.
(A) The deletion in the U3 region of the SIN LTR vector (lentiCRISPRv2) was repaired by inserting the full-length HIV-1LAI LTR sequence to create the HIV-CRISPR construct. (B) The ISG-targeting sgRNA library (15,348 unique sgRNA sequences) was synthesized and assembled into the HIV-CRISPR backbone to create the PIKAHIV (Packageable ISG Knockout Assembly) library. (C) THP-1 cells containing the PIKAHIV CRISPR knockout library were stimulated overnight with 1000 U/mL IFNα and infected with HIV-1LAI in duplicate infections. Viral RNA and genomic DNA were collected 3 days post infection and sgRNA sequences present in virions (vRNA) and genomic DNA (gDNA) were quantified through RT-PCR/PCR and deep sequencing. (D) Average enrichment of sgRNA sequences in the viral RNA (vRNA) was compared to their representation in the sequenced genomic DNA (gDNA) (n = 2). Y-Axis: Log2-normalized Fold Change (log2FC) of vRNA sgRNA sequences as compared to gDNA sgRNA sequences. X-Axis: random order of individual sgRNAs. The 200 non-targeting control (NTC) sgRNAs are shown in gray. The most enriched sgRNA sequences are in cyan (top 500), most depleted in orange (bottom 500). (E) MAGeCK Gene analysis was performed to identify the highest-scoring genes based on sgRNA frequencies for each gene across both replicates. An NTC gene set was created in silico by iteratively binning the 200 Non-Targeting Control sgRNA sequences into NTC Genes. Y-Axis: -log10MAGeCK Gene Score (positive). The type I IFN pathway genes IFNAR1, IRF9, STAT1 and STAT2 are shown in magenta. Non-Targeting Controls (NTCs) are in gray. Candidate Hits are cyan. The top 20-scoring genes across replicate screens are shown.
Figure 1—figure supplement 1. PIKAHIV ISG library gene and sgRNA selection.
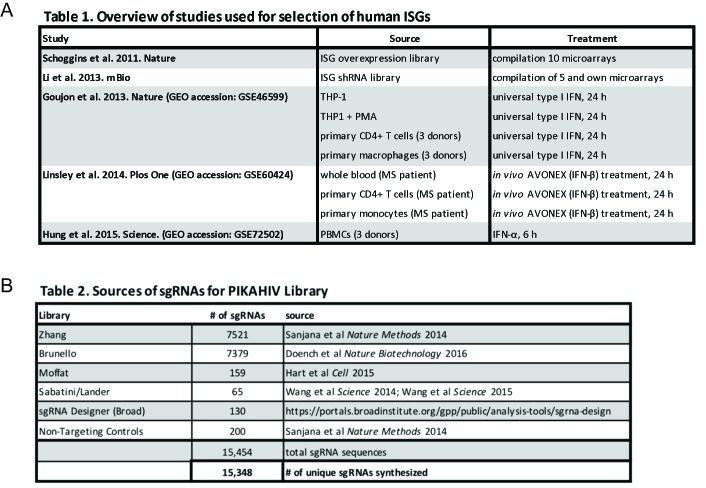
Figure 1—figure supplement 2. sgRNA read counts for the sequenced PIKAHIV Plasmid Library.