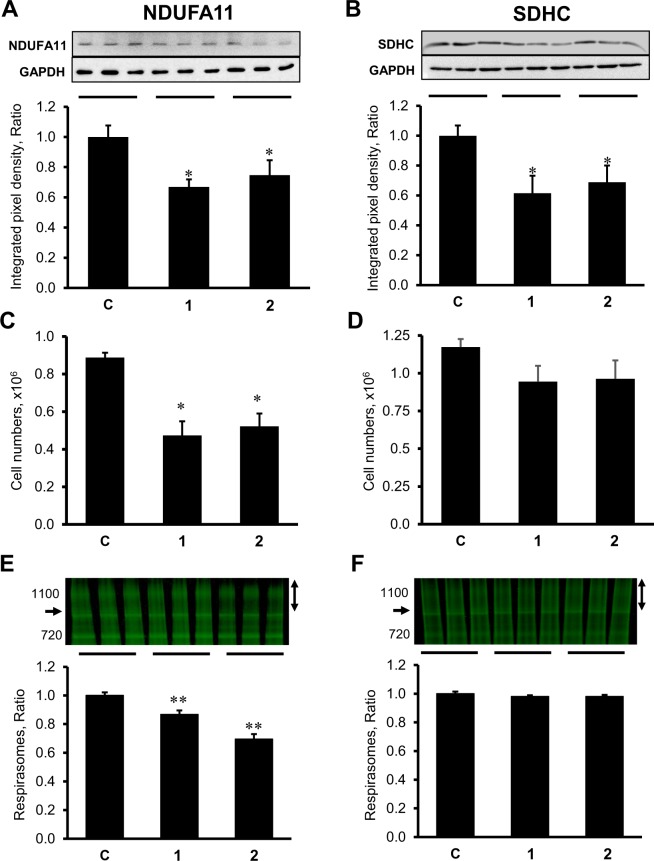
Figure 2.
The effects of NDUFA11 and SDHC silencing on protein expression levels, cell numbers and respirasome levels in H9c2 cells. (A,B), Protein levels of NDUFA11 and SDHC assessed by SDS-PAGE and Western blotting using specific antibodies against each protein. On the top of each graph, representative blots of NDUFA11 and SDHC are shown. (C,D), Cell numbers determined with the trypan blue exclusion assay and given in millions. (E,F), Quantitative data of respirasome levels. Respirasome levels were calculated as the ratio of integrated pixel densities from the bands containing complex I, III, and IV (marked as vertical double headed arrows in (E,F) to whole lane density. Data were normalized to control (non-treated) mitochondria. Arrows on the left side of gels indicating lowest molecular weight band containing complexes I, III, and IV. Identification of each bands were previously shown in13,14. Data were normalized to the control group. On the top of each graph, representative images of BN-PAGE gels are shown. The gels were stained by Coomassie brilliant blue G250. All parameters were measured 48 h after transfection. Two clones of siRNAs (1 and 2) per each gene have been used to silence NDUFA11 and SDHC. In control group (C), the cells were treated with negative control siRNA. Results are presented as mean ± SE. *P < 0.05, **P < 0.01 vs. control; n = 3 per group.