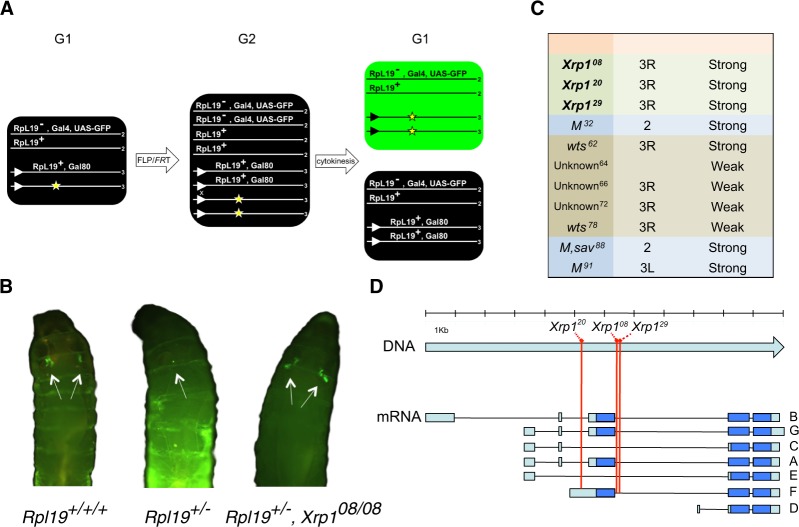
Figure 1.
Xrp1 mutations suppress cell competition driven elimination of loser cells in an EMS-based screen. Schematic of the genetics used to generate RpL19+/− loser clones in a wild-type background using the FLP/FRT system. Lines represent chromosomes, numbers at the end of each line indicate the chromosome number and triangles represent FRTs on the right arm of chromosome 3. Site-directed recombination between FRTs occurs when the expression of FLP is induced via heat shock. The yellow asterisk marks the chromosome to be tested for the presence of an EMS induced suppressor. The arrangement depicted here is a variation of the classical. MARCM technology that allows us to GFP label cells that are RpL19+/− and homozygous for a mutagenized chromosome arm 3R (A). Representative examples of living larvae displaying GFP clones in the pouch of the wing imaginal discs. SalE drives Gal4 expression in the wing pouch. (Left) Positive control for clone induction using the FRT82 RpL19+ chromosome. Recombination generates RpL19+/+/+ cells that are not eliminated. (Middle) Negative control for clone induction using the isogenized FRT82 chromosome. Recombination produces RpL19+/− cells that are efficiently eliminated. (Right) Suppressor Xrp108 rescues the elimination of RpL19+/− cells (B). List of suppressive mutations retrieved with the EMS screen. Intronic mutations Xrp108, Xrp120 and Xrp129 are strong suppressors (C). Different Xrp1 mRNA isoforms (from A to G). Blue color indicates the coding regions and light blue the untranslated regions. The red lines indicate the position of the three Xrp1 alleles retrieved from the EMS screen (Xrp108, Xrp120 and Xrp129) (D).