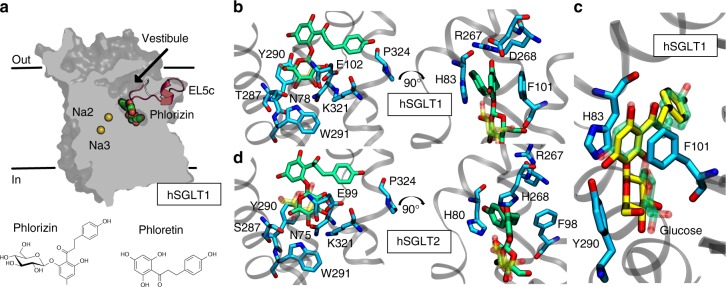
Fig. 1.
Predicted binding mode of phlorizin to hSGLT1 and hSGLT2. a Phlorizin bound to the outward-facing hSGLT1. The protein is gray, phlorizin is green and red, and Na+ at the putative Na2/Na3 sites are yellow. EL5c (red) is represented as cartoon. The 2D structures of phlorizin and phloretin are also shown. b Close-up view of phlorizin bound to hSGLT1. The sugar moiety occupies the sugar binding site aligning with the docked glucose (transparent yellow/red) where it makes excellent hydrogen bonding with the protein. The aglycon tail creates face-to-face aromatic π–π interactions with H83 and F101 and a cation–π interaction with R267 on EL5C. c Phloretin and glucose bound to hSGLT1. Phloretin (yellow/red) superposes over the aglycon tail from the phlorizin molecule (transparent green/red) in b, while glucose (yellow/red) aligns well with the sugar moiety. d Close-up view of phlorizin bound to hSGLT2. The binding mode is conserved in hSGLT2, although the aglycon tail is more tightly packed due to an aromatic cage that forms around the central ring made up of residues H80, F98, and H268 on EL5C