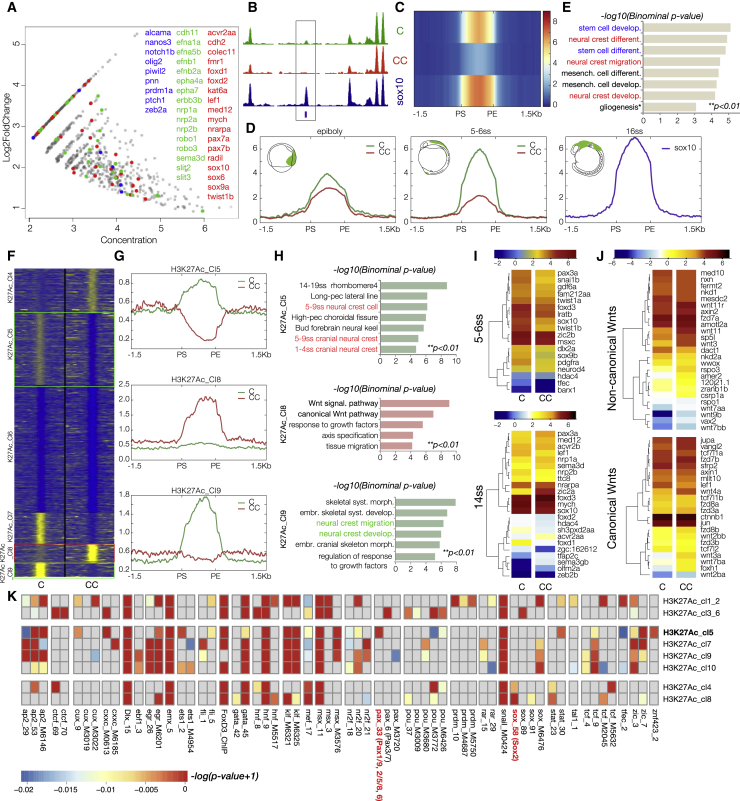
Figure 6.
Differential ATAC-Seq Analysis and Clustering of Enhancers Based on H3K27Ac Profiles
(A) Annotated MA plot depicting late opening enhancers significant by DiffBind analysis (p < 0.05, FDR<0.1) of the ATAC-seq signal at 5–6ss with annotated associated genes (stem cell genes, blue; cell adhesion/migration cues, green; NC specification and differentiation, red).
(B) Genome browser screenshot exemplifying the type of element isolated by DiffBind (boxed).
(C and D) (C) Heatmap (raw read counts) of all elements and (D) collapsed merged profiles indicating that identified elements are closed at epiboly and only start to open at 5–6ss.
(E) Functional annotation of DiffBind-identified enhancers shows association with later roles in NC (∗∗p < 0.01).
(F–H) (F) Heatmap depicting k-means linear enrichment clustering of H3K27Ac signal across non-promoter ATAC-seq peaks in foxd3-mutant (CC, Citrine/Cherry) and control (C, Citrine) at 5–6ss, (G) associated mean merged profiles for selected clusters, and (H) corresponding ontology enrichment bar plots indicating functional role of selected clusters.
(I) Heatmaps showing expression of NC specification genes (log FPKM) associated with K27Ac_Cl5 at 5–6ss and NC migration/differentiation genes associated with K27Ac_Cl9 at 14ss in foxd3-mutant (CC) and control cells (C).
(J) Heatmap depicting expression at 14ss of canonical and non-canonical Wnt pathway molecules (in log FPKM) associated with K27Ac_Cl8 that displays an increase in enhancer K27 acetylation in mutants.
(K) TF binding motif map representing significantly enriched TFBS for TF expressed at 5–6ss across different K27Ac-clusters. See Figure S5 for other K27Ac clusters and corresponding ontology enrichment bar plots.