Figure 7.
Putative Mechanisms of the Bimodal foxd3-Mediated NC Gene Regulation
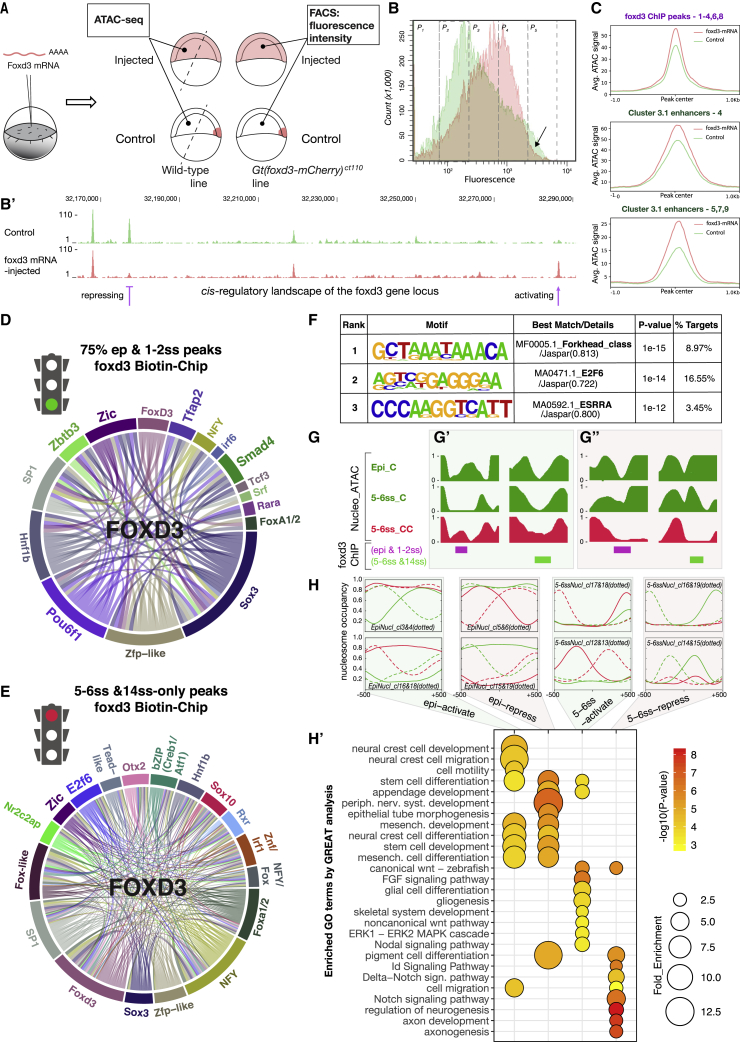
(A) Experimental strategy for foxd3 overexpression in vivo. Gt(foxd3-mCherry)ct110 heterozygous embryos were used for FACS and wild-type embryos for ATAC-seq experiments at 50% epiboly stages. For ATAC-seq, embryos were dissected (dashed lines) to only collect “foxd3-naive” cells that do not normally express foxd3. Native and ectopic foxd3 expression is illustrated in dark pink and lighter pink, respectively.
(B) FACS graph portraying a number of foxd3-mCherry expressing cells and underlying fluorescence intensities from control (green) and foxd3 mRNA injected (pink) embryos. P1–P5 – compartments of different fluorescence levels from the lowest to the highest. Black arrow indicates a loss of highest intensity fluorescence in foxd3 mRNA injected embryos versus control. (B′) Genome browser screenshot depicting region ∼60 kb upstream from the foxd3 transcription start site (TSS). Green and pink ATAC-seq tracks represent genome accessibility from control and foxd3 overexpressing embryonic cells. Purple arrows indicate either relative loss or acquisition of chromatin accessibility upon foxd3 overexpression.
(C) Mean density maps of merged profiles for k-means clusters featuring elements with differential accessibility between the foxd3 mRNA injected (in pink) and control (in green) 50% epiboly-staged embryos using either foxd3 binding maps or k-cluster 3.1 elements as a reference.
(D) Circle plot showing statistically significant TF motif co-occurrences on the “early NC” foxd3-bound activating elements.
(E) Circle plot showing different statistically significant TF motif co-occurrences on the “late NC” foxd3-bound elements, underlying repressive activity.
(F) De novo TF binding motifs enriched within foxd3-bound elements associated with NC genes negatively regulated by foxd3 at 14ss.
(G) Nucleosomal occupancy profiles expressed as relative NucleoATAC normalized cross-correlation signals. Profiles show changes in nucleosome positioning within the regulatory elements in control (C; green) and foxd3-mutant (CC; red) cells. Direct foxd3 binding at either early (epiboly, 1–2ss in magenta) or late stage (5–6ss, 14ss in green) results in either nucleosome clearing (G′; permissive role) or nucleosome compaction (G″; repressive role). Both processes are altered and nucleosomal patterns inverted in foxd3-mutant NC (G′ and G″).
(H) Mean density maps of merged profiles of nucleosomal clusters obtained by k-means analysis showing differential nucleosomal patterns between foxd3-mutants (CC; red) and controls (C; green). Both nucleosome-loose clusters of elements with activating patterns (epi-activate and 5–6ss-activate) and nucleosome-compact clusters with repressive patterns (epi-repress and 5–6ss-repress) are identified. (H′) Bubble chart depicting functional annotation of different nucleosomal clusters by GREAT (Bonferroni; p < 0.01). Only elements directly bound by foxd3 are analyzed.