Figure 2:
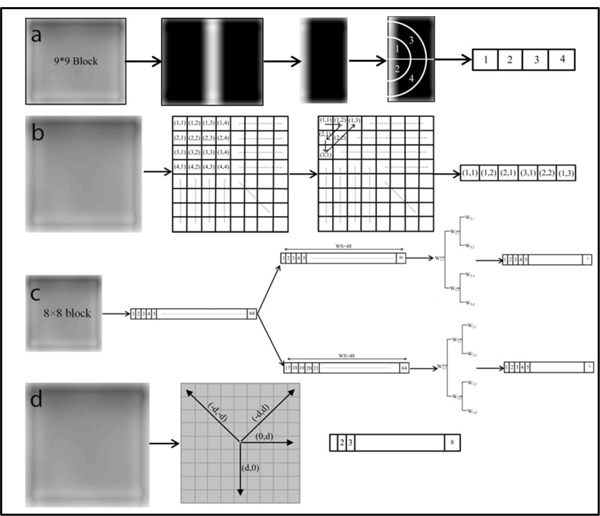
The feature array constructed for each block using the (a) FFT, (b) DCT, (c) Wavelet, and (d) GTDM. The process of establishing the four types of features can be summarized as follows. To start, each target slice was divided into either 8×8 or 9×9-pixel blocks (Figure 1), followed by image denoising. Each block was then processed with these four methods to generate the respective coefficient matrix. The representative part of the coefficient matrix for each block was determined accordingly (a-d) and arranged into a feature array. The feature arrays for all blocks were assembled to build a feature matrix that was directly used to compute the four types of features. For (a), we calculated the standard deviation of the FFT coefficients for the quarter annular regions to generate the feature array. In (b), six of upper left corner elements of the generated coefficient matrix were collected for each block and rearranged as a feature vector. As for (c), the pixel values of each 8 × 8-pixel block were rearranged as a 1-D vector that was further separated into two sequences, each of which contained 48 elements with 32 element overlap. The log root mean square (LRMS) value 9 was then calculated for the six sub-bands and the original sequence (i.e. W11) to establish 7 features elements for each sequence. For each 8 × 8-pixel block in (d), we evaluated the P function for eight gray levels in four different directions, leading to a total of 32 elements.
