FIGURE 1.
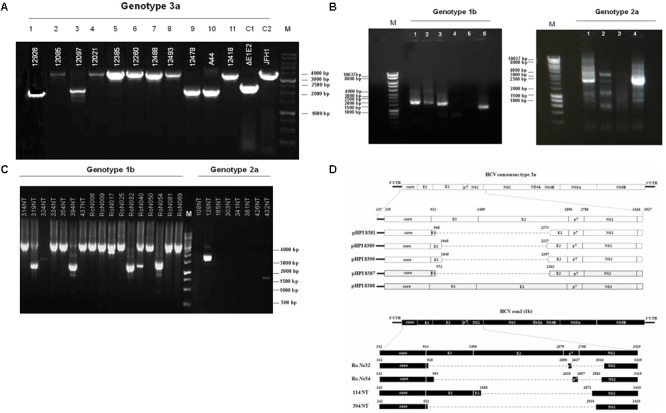
Detection of HCV natural mutants, containing large in-frame deletions in sera samples and liver tissue of HCV-infected individuals with different genotype. (A) Nested PCR amplicons from HCV 3a-infected sera samples were examined on 1% (w/v) agarose gel. Lanes 1, 3, 9, 10 correspond to sera samples containing IFDMs. The remaining lines 2, 4, 5, 6, 7, 8, 11 correspond to sera samples containing full length genomes. Positive controls are depicted in lanes C1, JFH1 full length; C2, JFH1 ΔE1E2. Line M, molecular size marker 1 Kb. (B) PCR amplicons from 1b and 2a HCV samples were similarly examined, using 1 Kb marker, where the IFDMs were observed on 1, 2, 3, 6 lanes for 1b and on 1, 2, 4 lanes for 2a. (C) Nested-PCR amplification products from liver tissue samples of HCV-infected patients (genotypes 1b and 2a). (D) Schematic representation of the defective and full length genomes’ architecture identified in the sera samples and liver tissues of HCV patients chronically infected with genotypes 3a and 1b.
