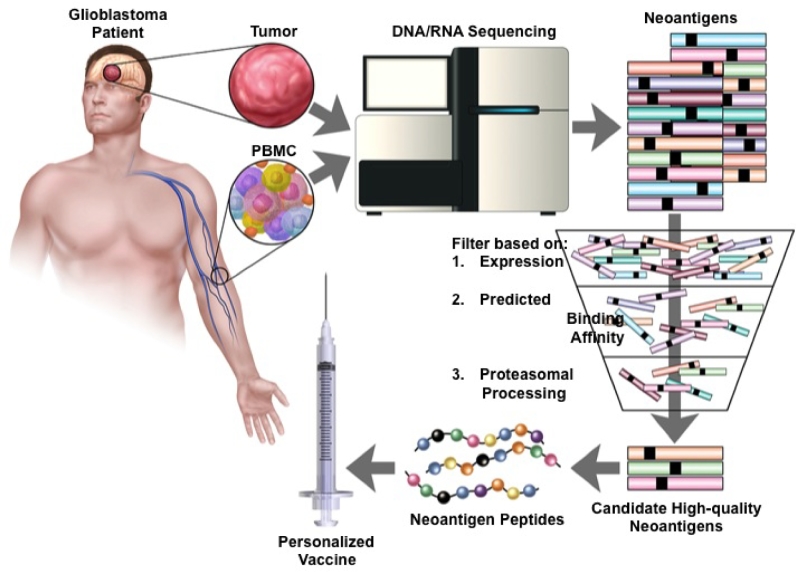
FIGURE 3.
Schematic representation of cancer immunogenomics workflow for neoantigen discovery. Normal reference tissue (ie, PBMC) and tumor tissue is obtained and undergoes DNA whole exome and RNA sequencing to identify somatic, nonsynonymous mutations. Tumor-specific mutations are then filtered using computational software to prioritize neoantigens based on expression, predicted patient-specific HLA binding affinity, and likelihood of endogenous proteosomal processing. Peptides corresponding to candidate high-quality neoantigens are then manufactured and administered back to the patient as a personalized vaccine. PBMC, peripheral blood mononuclear cell.