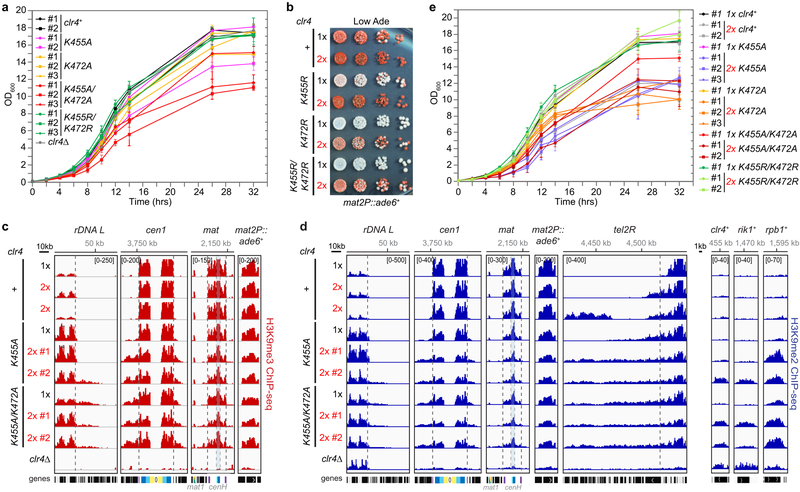
Extended Data Figure 7 ∣. Disruption of Clr4 auto-inhibition results in growth defects, inappropriate H3K9me spreading and formation of new H3K9me domains.
a, Growth assays showing that cells expressing the indicated Clr4 hyperactive mutants have growth defects. Data are means ± s.d. from three biological replicates. b, Silencing assay of mat2P::ade6+ on low-adenine medium (Low Ade) with one (1x) or two (2x) copies of clr4+ or mutant clr4 genes. See main text for details. Experiment performed twice with at least two independent clones with similar results. c-d, H3K9me3 (c) or H3K9me2 (d) ChIP-seq reads mapped to different heterochromatin regions (left) or euchromatic genes (right) in the indicated genotypes are presented as reads per million (number in bracket in the first row of each ChIP-seq data). Top, chromosome coordinates. See Fig. 3 legend for abbreviations. Sequencing performed once with two independent clones as shown. (e) Growth assays showing that the addition of a second copy (2x) of the clr4 gene encoding hyperactive Clr4 mutants exacerbate growth defects. Data are means ± s.d. from three biological replicates.