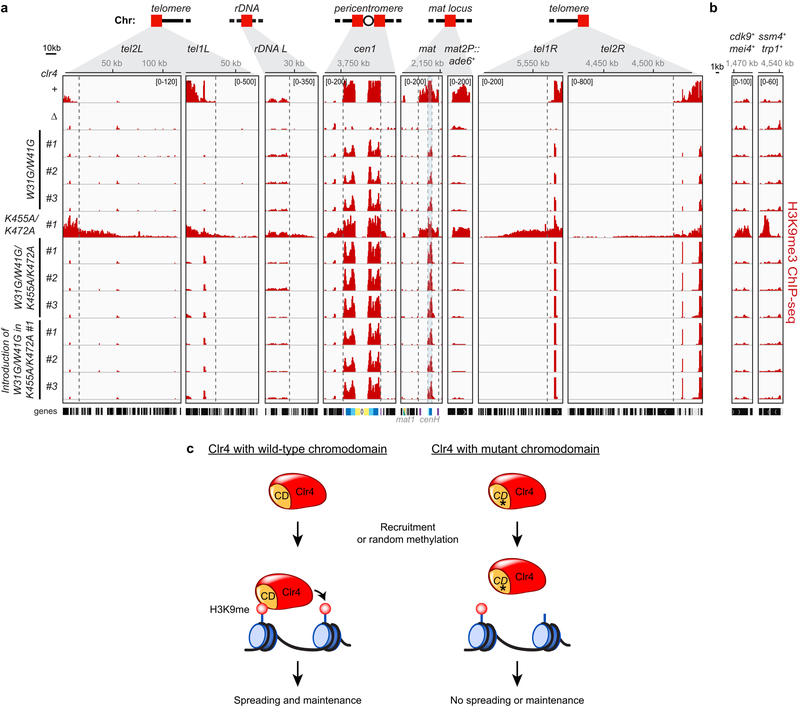
Extended Data Figure 9 ∣. Clr4 autoregulation prevents illegitimate heterochromatin formation mediated by the Clr4 read-write positive feedback mechanism.
a-b, H3K9me3 ChIP-seq reads mapped to different heterochromatin regions (a) or euchromatic genes (b) in the indicated genotypes are presented as reads per million (number in bracket in the first row of each ChIP-seq data). Top, chromosome coordinates. See Fig. 3 legend for abbreviations. Sequencing performed once with the indicated independent clones (#) for each genotype. c, Schematic summary showing Clr4 chromodomain (CD)-dependent spreading and maintenance of histone H3K9me. The CD-dependent positive feedback loop is critical even with hyperactive Clr4.