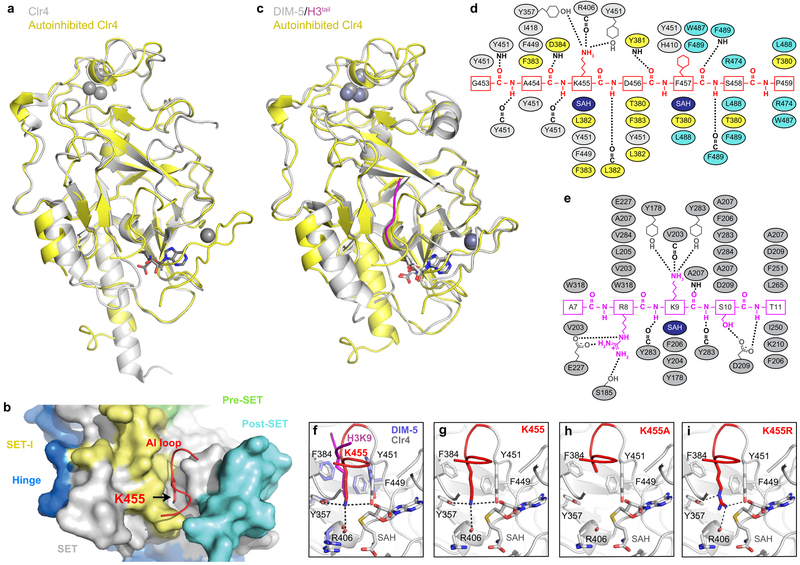
Extended Data Figure 3 ∣. Comparison of Clr4 structure with other SUV39H methyltransferases.
a, Alignment of the autoinhibited Clr4192–490 (yellow) and the previously determined Clr4192–490 structure lacking post-SET domain and cofactor (grey) (PDB ID 1MVH). b, Close up view of S. pombe Clr4192–490 autoinhibited conformation (surface) showing that K455 (red stick) in the AI loop (red) is located within the active site pocket. c, Alignment of autoinhibited Clr4192–490 (yellow) and DIM-5 (grey) in complex with histone H3 peptide (magenta) and SAH (PDB ID 1PEG). d-e, Schematic representation of residues that engage in interactions between Clr4 (grey, yellow and cyan) and Clr4 AI loop (red) (d) and DIM-5 (grey) and histone H3 (magenta) (e). Hydrogen bonds and salt bridge interactions are shown as dashed lines. Color assignments are as in Fig. 1d; residues in the SET-I, SET, and post-SET domain are shown in yellow, grey and cyan, respectively. SAH is shown in dark blue. f, Alignment of the catalytic pockets of Clr4192–490 and DIM-5 (PDB ID 1PEG) showing that Clr4-K455 (red) and lysine 9 of the histone H3 substrate (magenta) occupy similar positions. g-i, Close up view of the autoinhibited conformation of Clr4 active site (g, as in Fig. 1h, shown here for comparison) and modeling the K455A (h) and K455R (i) mutants in the active site. Possible bonding interactions are shown as dashed lines.