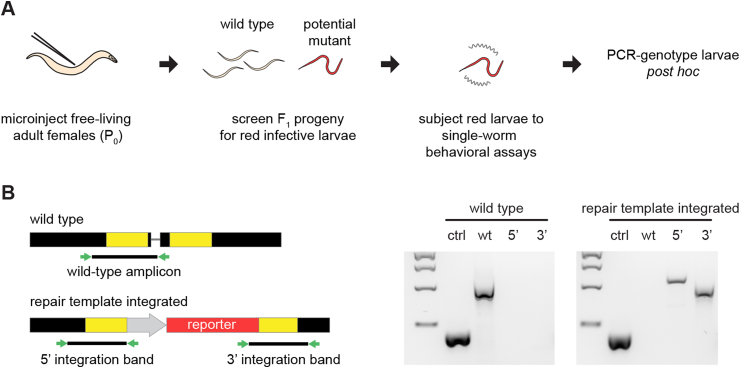
Fig. 4.
A pipeline for CRISPR/Cas9-mediated targeted mutagenesis in S. stercoralis. A. DNA plasmids containing the CRISPR components (the Cas9 gene, a single guide RNA, and a repair template containing a red fluorescent marker for homology-directed repair) are introduced into free-living adult females (P0) by gonadal microinjection. The F1 iL3 progeny are then screened for the presence of the fluorescent marker. Individual F1 iL3s expressing the fluorescent marker are subjected to single-worm behavioral or physiological assays, and then PCR-genotyped post hoc. B. Individual F1 iL3s are PCR-genotyped for homozygous disruption of the gene of interest and integration of the repair template (Gang et al., 2017; Bryant et al., 2018). Left, diagram depicting the expected PCR products from single-worm genotyping. Approximate locations of primer binding sites are shown as green arrows. In the diagram of the wild-type chromosomal locus (top), the thin grey line represents the location of the CRISPR target site and the yellow regions indicate the DNA sequences that match the homology arms of the repair template. In the diagram of the chromosomal locus after the repair template has integrated, the grey arrow indicates the promoter used to drive expression of the fluorescent reporter gene and the red region indicates the coding sequence of the reporter gene. The wild-type amplicon will yield a PCR product only if the unedited wild-type locus is present. The 5′ integration band tests for integration of the 5′ end of the repair template; the 3′ integration band tests for integration of the 3′ end of the repair template. For both PCR reactions testing for integration of the repair template, one primer matches a sequence in the repair template and the other matches a sequence in the flanking genomic DNA. Thus, these reactions will only yield a PCR product following successful homology-directed repair. Right, examples from DNA gels showing the PCR products obtained from a wild-type iL3 and a repair-template-integrated iL3. ctrl = a positive control that amplifies a wild-type region of the S. stercoralis actin-2 gene; wt = wild-type amplicon; 5’ = 5′ integration band; 3’ = 3′ integration band. DNA ladder shows 2 kb, 1.5 kb, 1 kb, and 500 bp bands from top to bottom. Figure adapted from Gang et al. (2017). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)