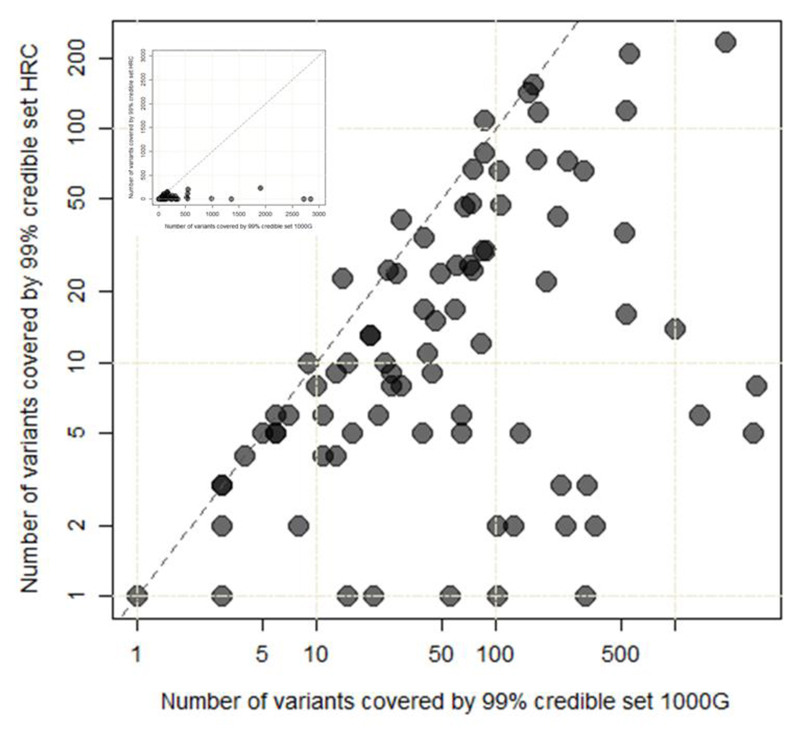
Figure 4. Comparison of fine-mapping resolution at 83 distinct signals.
The number of variants included in the 99% credible set for each of the 83 distinct signals constructed using meta-analysis of GWAS data imputed using the 1000G multi-ethnic reference panel (26,676 T2D cases and 132,532 controls) (x-axis; logarithmic scale) is plotted against those (y-axis; logarithmic scale) derived using HRC-based imputation (74,124 T2D cases and 824,006 controls). Inset presents the same plot but with linear scales.