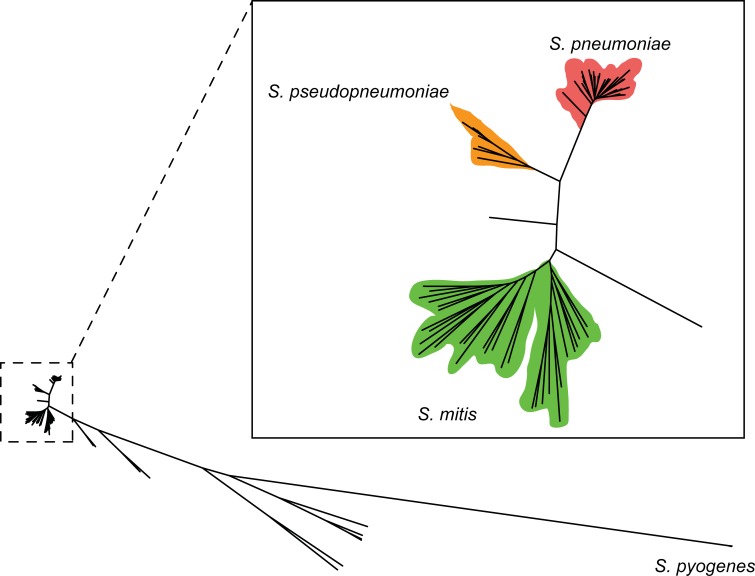
Fig 1. Phylogenetic tree based on core genome analysis of the strains included in the “Curated Database”.
The tree includes all strains of S. pneumoniae, S. pseudopneumoniae, S. mitis, including the experimental strains used in the study, as well as the type strain of the other species of the Streptococcus Mitis Group included in the database and S. pyogenes. The tree is based on 168,439 homologous amino acid positions and was constructed, using PhyML software and the aLRT algorithm.