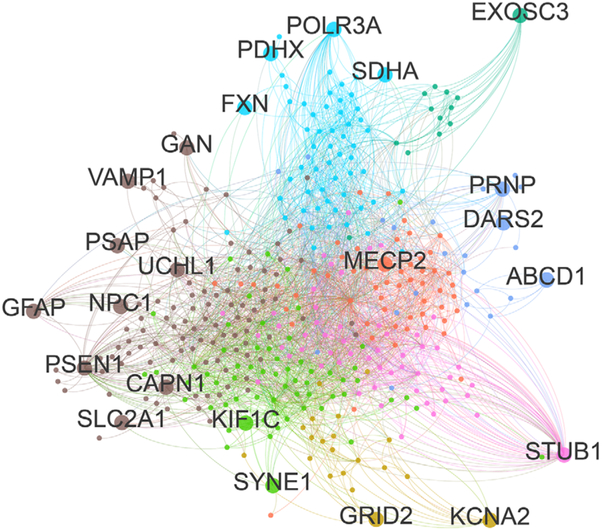
FIG. 2.
Protein-protein-interaction network for ataxia-HSP spectrum disease genes. To generate a protein-protein-interaction (PPI) network, interactors (iteration 1) of 63 spastic ataxia genes listed in Table 1 were extracted using the iRefScape plugin in Cytoscape v2.8.3.60-62 Autosomal-dominant repeat-expansion genes were removed from the seed list, as their binding properties appear to be largely shaped by their polyglutamine tracts rather than the properties of the wild-type protein. The PPI network was then imported into Gephi v0.9.1 and filtered, whereby nonhuman interactions, predicted interactions, interactions based solely on high-throughput experiments, self-loops, and nodes with a degree < 2 were removed, retaining only nodes that were maximum 1 degree removed from the input spastic ataxia seed genes. The resulting network contained 389 nodes and 2582 undirected edges. We then applied the Louvain method to detect communities, neighborhoods of highly connected nodes. A total of 8 communities were detected, represented by differently colored nodes and edges in the figure. Ataxia-spasticity spectrum seed genes are represented by larger dots and labeled with the respective gene name. [Color figure can be viewed at wileyonlinelibrary.com]