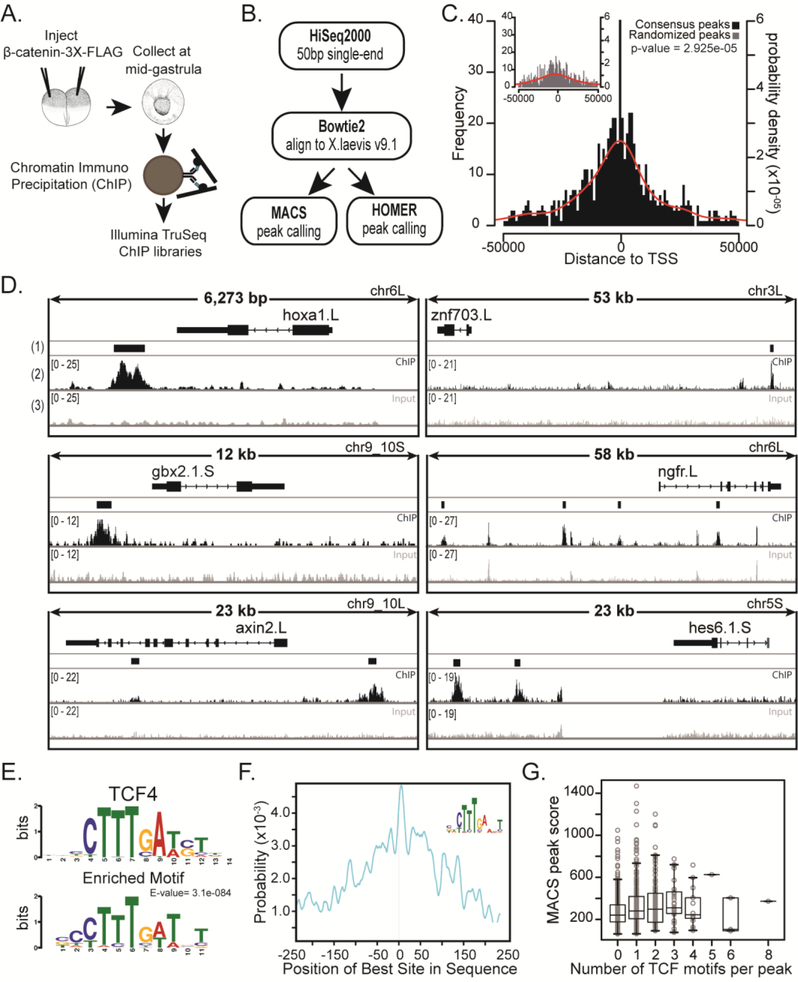
Figure 2. Chromatin Immunoprecipitation and sequencing (ChIPseq) of a FLAG-tagged β-catenin at mid gastrula identifies β-catenin bound regions.
(A) Schematic of experiment. 500 pg of mRNA encoding triple FLAG-tagged β-catenin was injected animally into both blastomeres of a 2-cell stage embryo. Chromatin was crosslinked at mid gastrula, sonicated, and used to make Illumina TruSeq libraries. (B) Pipeline of ChIPseq analysis. 50 bp single-end reads were aligned to X. laevis genome version 9.1 using Bowtie2. Peaks were called with MACS and HOMER. Common peaks were identified by both peak callers in all three biological replicates. (C) Histogram of distances from peak to transcription start site (TSS). Inset: Common peaks were randomly distributed along the genome to make Randomized Peaks (see methods). Red line is the probability density for distances. For ChIP peaks the mean distance to TSS = - −2929 bp and for randomized peaks the mean distance to TSS = −9131 bp. Samples are significantly different with a p-value = 2.925e-05 (Kolmogorov-Smirnov test). Note: Only peaks within 50kb from TSS are shown. (D) IGV browser views of ChIPseq coverage at previously identified target genes and new candidate targets. (1) The Consensus Peak track shows the consensus peaks from three replicate experiments. (2) Read pile-up coverage from a single ChIPseq replicate sample. We note that the width of the peak displayed depends on the overall length of DNA represented, such that peaks appear narrow in 50kb windows. (3) Read pile-up coverage from a single Input replicate sample. (E) Peaks were extended by 250bp from the middle nucleotide and submitted to MEMEchip. Position Weight Matrix of TCF4 motif and most enriched motif found in consensus peaks. E-value=3.1e-084. (F) Distribution of enriched motif in consensus peaks. (G) Box plot representing number of TCF4 motifs in a peak versus the MACS score (MACS score = −10*log10pvalue) of that peak.