ABSTRACT
Activator of G-protein signaling 3 (AGS3, also known as GPSM1) exhibits broad functional diversity and oscillates among different subcellular compartments in a regulated manner. AGS3 consists of a tetratricopeptide repeat (TPR) domain and a G-protein regulatory (GPR) domain. Here, we tested the hypothesis that phosphorylation of the AGS3 GPR domain regulates its subcellular distribution and functionality. In contrast to the cortical and/or diffuse non-homogeneous distribution of wild-type (WT) AGS3, an AGS3 construct lacking all 24 potential phosphorylation sites in the GPR domain localized to cytosolic puncta. This change in localization was revealed to be dependent upon phosphorylation of a single threonine amino acid (T602). The punctate distribution of AGS3-T602A was rescued by co-expression of Gαi and Gαo but not Gαs or Gαq. Following treatment with alkaline phosphatase, both AGS3-T602A and WT AGS3 exhibited a gel shift in SDS-PAGE as compared to untreated WT AGS3, consistent with a loss of protein phosphorylation. The punctate distribution of AGS3-T602A was lost in an AGS3-A602T conversion mutant, but was still present upon T602 mutation to glutamate or aspartate. These results implicate dynamic phosphorylation as a discrete mechanism to regulate the subcellular distribution of AGS3 and associated functionality.
KEY WORDS: Activators of G-protein signaling 3, G-protein regulatory domain, Cytosolic puncta, Phosphorylation, Phase transition, Aggresome
Summary: A single threonine residue in the G-protein regulatory domain of AGS3 is a determinant factor in the oscillation of AGS3 among different subcellular compartments and protein phase transitions.
INTRODUCTION
Activators of G-protein signaling (AGS) are a broad panel of biological regulators that influence signal transfer from receptor to G-protein, guanine nucleotide binding and hydrolysis, G-protein subunit interactions, and/or serve as alternative binding partners for Gα and Gβγ independently of the classic heterotrimeric Gαβγ for specific G-protein subtypes (Blumer and Lanier, 2014; Blumer et al., 2007, 2012). Such proteins fall into four general groups (Groups I–IV) based upon their biochemical and cell-based interaction with G-protein subunits, and exhibit a wide range of functionality related to basic cellular processes and their dysregulation in various disease states. Group II AGS proteins [AGS3 (also known as GPSM1), AGS4 (GPSM3), AGS5 (LGN or GPSM2), PCP2 (L7 or GPSM4), AGS6 (RGS12) and RGS14] contain 1–4 G-protein regulatory (GPR) motifs that interact with Gαi, Gαt and Gαo, and stabilize the GαGDP complex free of Gβγ (Blumer and Lanier, 2014; Blumer et al., 2007, 2012). AGS3 and AGS5 are both modular proteins, comprising N-terminal tetratricopeptide repeats (TPR) and four G-protein regulatory motifs (GPR), with the TPR and the GPR domains connected by a linker region. Although it has a similar domain organization to AGS5, AGS3 has a broader functional portfolio and the two GPR proteins exhibit different subcellular distributions and functions. AGS5 plays a key role in spindle orientation during asymmetric cell division as well as in planar cell polarity, and mutations in the protein cause specific types of hearing loss due to disorganization in the cochlear cell cilia structure (Mauriac et al., 2017; Saadaoui et al., 2017).
Among Group II AGS family members, AGS3 exhibits the greatest functional diversity and is implicated in a wide range of system and cellular functions, including adaptive responses to addiction and craving behavior, autophagy, membrane protein trafficking, metabolism, cardiovascular function, the renal response to injury (cytoprotection), ciliary biology, cystogenesis, polycystic kidney disease, asymmetric cell divisions and immune responsiveness (Blumer and Lanier, 2014; Blumer et al., 2012, 2008; Branham-O'Connor et al., 2014; Robichaux et al., 2017; Vural et al., 2016, 2010; Bowers et al., 2004; Groves et al., 2010; Kwon et al., 2012; Regner et al., 2011; Sanada and Tsai, 2005; Saadaoui et al., 2017; Chishiki et al., 2017; Choi et al., 2016; Pedram et al., 2017; Singh et al., 2014; Yeh et al., 2013). How such functional diversity is achieved is not known, but may relate to the impact of the protein on basic cellular processes in a cell- and system-type specific manner.
AGS3 also oscillates among different subcellular compartments in a regulated manner and its expression is cell-type specific, developmentally regulated, and upregulated in response to cellular stress; all of which are likely intimately related to the functional diversity of the protein (Blumer and Lanier, 2014; Blumer et al., 2012; Vural et al., 2010; An et al., 2008; Oner et al., 2010, 2013; Garcia-Marcos et al., 2011; Groves et al., 2007; Pattingre et al., 2003; Xu et al., 2010; Regner et al., 2011; Kwon et al., 2012). Two major questions still to be answered are, firstly, what controls the subcellular distribution of AGS3 and, secondly, how is the regulated movement of the protein within the cell involved in AGS3 functionality? Cellular stress or disruption of the TPR organizational structure of AGS3 results in the localization of AGS3 to punctate subcellular structures distinct from defined intracellular organelles or vesicles. Further, this localization is regulated by interaction with AGS3 binding partners Gαi and the cell polarity protein inscuteable (Vural et al., 2010). Moreover, activation of certain G-protein-coupled receptors results in the dissociation of an AGS3–Gαi signaling complex, releasing AGS3 from the cell cortex into the cytosol with a subsequent enrichment of AGS3 at the trans-Golgi network of the Golgi apparatus and Golgi fragmentation (Oner et al., 2010, 2013; Robichaux et al., 2015). It is suggested that the observed range of subcellular distribution of AGS3 actually reflects a more dynamic movement of the protein within the cell with its presence in any given microdomain stabilized by regulated protein conformation, binding partners and/or transient posttranslational modification.
The subcellular distribution of AGS3 into subcellular puncta and its regulation is of particular interest. The GPR region of AGS3 is a key regulatory domain for both functional impact and the distribution of AGS3 within the cell, and the linker and/or GPR domains are postulated to be sites for regulated protein phosphorylation of Group II AGS proteins (Blumer et al., 2003; Fukukawa et al., 2010; Hao et al., 2010; Groves et al., 2010). Toward this end, we focused on the role of the GPR domain in determining the distribution of the protein into the puncta landscape. As described here, elimination of 24 candidate serine and threonine phosphorylation sites in the GPR domain of AGS3 resulted in robust formation of cytosolic, non-nuclear puncta, a gel shift of the protein as determined by SDS-PAGE, and a phase transition of the protein. We further report the identification of a single AGS3 residue (T602) as the key determinant of AGS3 distribution into the punctate structures. The distribution of the phosphodeficient protein into puncta was rescued by Gαi and Gαo but not Gαs or Gαq. Overall, the results implicate dynamic phosphorylation as a discrete mechanism to regulate the subcellular distribution of AGS3 and associated functionality.
RESULTS
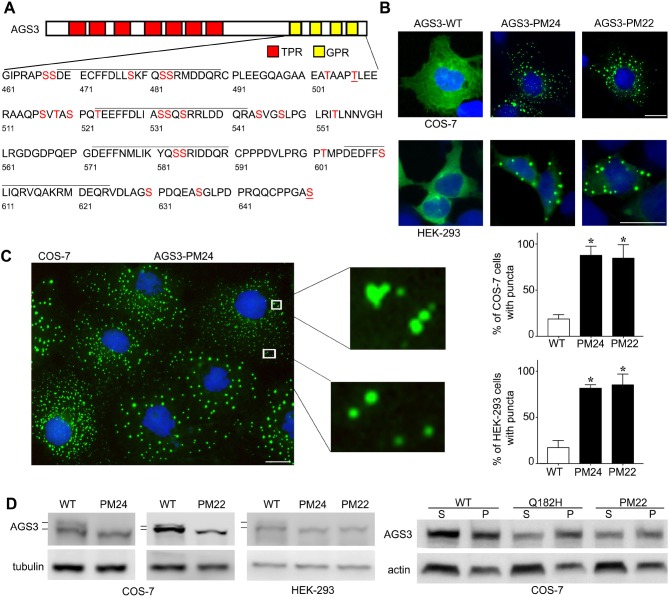
We first examined the subcellular distribution of AGS3 after all 24 of the candidate serine and threonine phosphorylation sites in the GPR domain were substituted with alanine [phosphomutant (PM) 24; Fig. 1A]. Surprisingly, in both COS-7 and HEK-293 cells, AGS3-PM24 assembled in cytosolic, extranuclear puncta (Fig. 1B,C). These puncta were visually similar to puncta we have previously observed containing AGS3-Q182H (single residue substitution in the TPR domain; Vural et al., 2010) and also to those containing the signaling protein Dishevelled (which has three homologs, DVL1–DVL3, in mammals) (Schwarz-Romond et al., 2007; Smalley et al., 2005; Bernatík et al., 2014; Greer et al., 2013; González-Sancho et al., 2013; Mlodzik, 2016; Vural et al., 2010). A punctate distribution of endogenous AGS3 is also apparent in renal epithelial Balb/c polycystic kidney (BPK) mice where AGS3 is upregulated (Nadella et al., 2010), but such a distribution is less well-defined in primary cultures of hippocampal neurons or in PC-12 neuronal cells (Blumer et al., 2002). In COS-7 cells, the percentage of AGS3-expressing cells with puncta in experiments with AGS3-PM24 (87.7±5.8%) and AGS3-PM22 (84.7±8.4%) was significantly increased above the percentage observed with WT AGS3 (18.7±2.7%) (Fig. 1C). Similar results were obtained in HEK-293 cells (WT AGS3, 17.3±4.4%; AGS3-PM24, 81.7±2.2%; AGS3-PM22, 85.3±6.7%).
Fig. 1.
Subcellular distribution and biochemical analysis of candidate phosphorylation sites in the G-protein regulatory domain of AGS3. (A) Schematic representation of AGS3 domain organization with sequence information for the GPR domain. The lines over the sequence indicate the four GPR motifs in the protein. The serine (S) and threonine (T) residues indicated in red were substituted with alanine in AGS3-phosphomutant (AGS3-PM) proteins. (B,C) Subcellular distribution of WT AGS3, AGS3-PM24 and AGS3-PM22 in COS-7 and HEK-293 cells. The images are representative of five separate experiments and are shown at a magnification of 40×. Scale bars: 10 µm. Graphs in C show the percentage (mean±s.e.m.) of COS-7 and HEK-293 cells exhibiting cellular puncta. 200 cells were counted for each independent transfection (n=5). *P<0.05 by one-way ANOVA followed by Tukey's multiple comparison test. (D) Left: Immunoblotting of cell lysates from COS-7 and HEK-293 cells expressing WT AGS3, AGS3-PM24 or AGS3-PM22. Right: Immunoblotting of fractionated cell lysates from COS-7 cells expressing WT AGS3, AGS3-Q182H or AGS3-PM22, showing results from supernatant (S) and pellet (P) fraction samples. The images shown are representative of one (right panel) to five (left panel) separate experiments.
Denaturing polyacrylamide gel electrophoresis (SDS-PAGE) revealed a migration shift of AGS3-PM24 as compared to WT AGS3, consistent with a loss of protein phosphorylation (Fig. 1D) (Groves et al., 2010). WT AGS3 appears as multiple species by SDS-PAGE and immunoblotting and AGS3-PM24 exhibits a faster migration and a loss of one of the slower migrating species observed for WT AGS3 (Fig. 1D, left panels). Similar results were obtained with AGS3-PM22, which has all of the alanine-substituted GPR serine and threonine residues in AGS3-PM24 with the exception of T507 and S650 (Fig. 1B–D). Subcellular fractionation followed by SDS-PAGE and immunoblotting indicated that, similar to AGS3-Q182H (Vural et al., 2010), the appearance of the puncta with the AGS3-PM22 phosphomutant was also associated with a phase transition from the soluble to pellet fractions as compared to WT AGS3 (Fig. 1D, right panel).
Identification of key serine and threonine residues leading to formation of AGS3-PM puncta
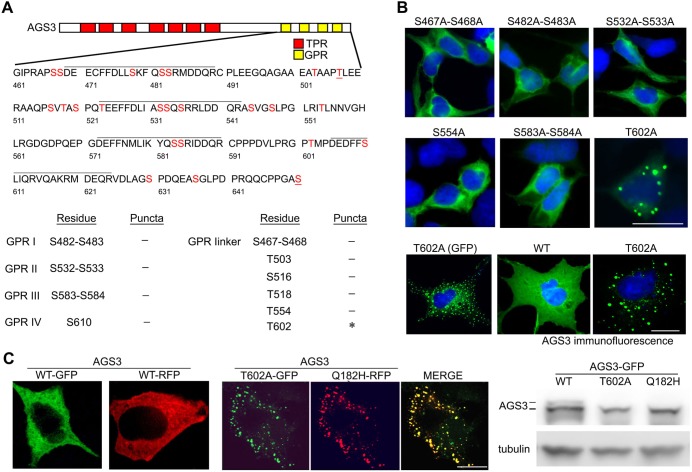
Of the 24 potential serine and threonine phosphorylation sites in the AGS3 GPR domain, six residues (S478, S482, S483, S516, S584, T602, S610) are conserved in the AGS3 orthologs from Homo sapiens, Rattus norvegicus, Mus musculus, Xenopus laevis and Drosophila melanogaster. We developed a mutational strategy to identify which of the 24 candidate phosphorylation serine and threonine residues contributed to the redistribution of AGS3 into the puncta. In the first set of experiments, we targeted amino acids in each of the four AGS3 GPR domains with the following substitutions: GPR I, S482 and S483; GPR II, S532 and S533; GPR III, S583 and S584; GPR IV, S610. In the second set of experiments we targeted the linker regions around the individual GPR domains: Pre-GPR I, S467 and S468; GPR I–II, T503, S516 and T518; GPR II–III, T554; GPR III–IV, T602. Out of this series of constructs only AGS3-T602A generated the AGS3 puncta (Fig. 2A,B), suggesting that T602 was a critical determinant of the subcellular location of the protein. The punctate distribution of AGS3-T602A was also observed with non-GFP-tagged AGS3 (Fig. 2B). To further confirm the role of T602A as a determinant of the punctate distribution of AGS3, we examined the subcellular distribution of various other constructs with T602A present in different contexts. This series of experiments included the double mutant AGS3-S554A-T602A and three constructs with a varying number of alanine substitutions (AGS3-PM10, AGS3-PM13A, AGS3-PM13B). AGS3-PM13A and AGS3-PM13B differ only in alanine substitution for the last two serine residues (S630 vs S650). Each of the four constructs exhibited a punctate distribution of AGS3 (data not shown).
Fig. 2.
Determination of specific candidate serine/threonine phosphorylation site(s) in the subcellular distribution of AGS3. (A) Schematic representation of AGS3 domain organization. Serine (S) and threonine (T) residues indicated in the lower portion of the schematic were substituted with alanine. (B) Subcellular distribution of AGS3 in HEK-293 cells (upper and middle panel) or COS-7 cells (bottom panel) transfected with pEGFP::AGS3 constructs (250 ng). (C) Subcellular distribution and gel migration pattern of WT AGS3, AGS3-T602A and AGS3-Q182H in HeLa cells. Left panel, cells were transfected with WT pEGFP::AGS3 or WT pRFP::AGS3. Middle panel, cells were co-transfected with pEGFP::AGS3-T602A and pRFP::AGS3-Q182H constructs as indicated. Right, immunoblots of cell lysates from HeLa cells expressing WT AGS3, AGS3-T602A or AGS3-Q182H. Images in B,C are representative of three independent experiments, at 40× magnification. Scale bars: 10 µm.
Analysis of the AGS3 puncta
The number and size of puncta are cell-type specific and likely relate to differences between the cell types in cytosolic volume, AGS3 expression and/or specific biological processes related to protein processing and signaling dynamics. COS-7 cells exhibit a larger number of puncta (>50) and with a diameter of ∼0.25 to ∼1.6 µm for both AGS3-Q182H and AGS3-T602A as compared to those observed in HEK-293 cells (<20 puncta per cytosol; ∼0.3 to ∼1.1 µm diameter). In both cell types, the smaller puncta were well-rounded and enriched toward the cell cortex, whereas the larger puncta tended to be more irregular in shape and localized in the cytosol domains more toward the cell interior or nucleus (Fig. 1C). As noted earlier, AGS3 puncta, like dishevelled puncta, do not associate with defined subcellular vesicles or domains and also do not readily associate with the autophagy pathway (Vural et al., 2010, 2013; Pattingre et al., 2003). The subcellular distribution of WT AGS3 in COS-7 and HEK-293 cells was not altered by blockade of autophagic vesicle formation by inhibitors of class III phosphatidylinositol-3-kinase, by inhibition of mTOR kinase or by induction of the autophagy-lysosomal pathway by nutrient deprivation (data not shown).
AGS3 puncta observed with single residue substitutions in the TPR (Q182H) or GPR (T602A) exhibit remarkably similar size, shape and cellular distribution (Vural et al., 2010). The Q182 residue (Rattus norvegicus as used here; Q185 in Homo sapiens) was previously identified as a nonsynonymous AGS3 SNP (rs28507185) adjacent to the fourth TPR motif (Vural et al., 2010). We thus asked whether AGS3-PM and AGS3-Q182H were colocalized in the same puncta population and whether AGS3-PM puncta exhibited the same regulatory properties as AGS3 with the TPR point mutation. Co-expression of pEGFP::AGS3-T602A and pRFP::AGS3-Q182H in COS-7 cells followed by fluorescence cell imaging revealed that the two proteins were colocalized in the same cytosolic, nonnuclear puncta population (Fig. 2C, middle panel). SDS-PAGE and immunoblotting indicated that the two variants (TPR, AGS3-Q182H; GPR, AGS3-T602A), as compared to WT AGS3, also exhibited a similar migration pattern by SDS-PAGE with the loss of the slower migrating, putative phosphorylated species of AGS3 (Fig. 2C, right panel). These data suggest that introduction of the Q182H mutation results in an AGS3 conformation that impedes AGS3 phosphorylation and thus leads to the stabilization of AGS3-Q182H in a population of puncta similar to those observed with the phosphodeficient AGS3.
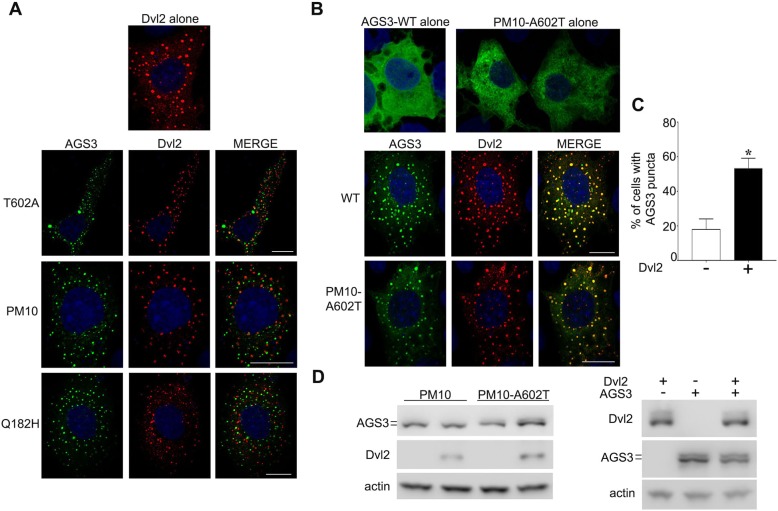
We also explored the relationship between the punctate structures observed for the TPR mutant (AGS3-Q182H) and the GPR mutant (AGS3-T602A) with punctate structures defined by other signaling proteins. Based on the apparent visual similarities in the punctate patterns of dishevelled proteins and the AGS3 variants, we asked whether AGS3 constructs were distributed into the same population of puncta as those containing DVL2 in COS-7 cells. As reported previously (Schwarz-Romond et al., 2007; Smalley et al., 2005; Bernatík et al., 2014; Greer et al., 2013; González-Sancho et al., 2013; Mlodzik, 2016; Vural et al., 2010), DVL2 was localized to punctate structures following expression in COS-7 cells (Fig. 3) and the DVL2 punctate structures were not altered by the expression of different AGS3 constructs (Fig. 3A,B). Co-expression of DVL2 with the phosphodeficient AGS3 constructs indicated that DVL2 and AGS3 variants localize to distinct punctate populations in the cell (Fig. 3A). However, WT AGS3 and AGS3-PM10-A602T, which typically exhibit a diffuse, non-homogeneous distribution in the cell, were actually redistributed into DVL2 punctate structures when coexpressed with DVL2 (Fig. 3B). The percentage of WT AGS3 puncta alone in the absence of DVL2 co-expression (18±3.5%) was significantly lower than the percentage of AGS3-T602A puncta observed with DVL2 co-expression (53.3±3.4%) (Fig. 3C). In contrast to the gel migration pattern observed with the AGS3-PM10 mutant, immunoblots of lysates from cells expressing the AGS3 conversion mutant AGS3-PM10-A602T indicated a change in the SDS-PAGE migration pattern with the appearance of a slower migrating species when coexpressed with DVL2 (Fig. 3D). The gel migration pattern exhibited with WT AGS3 was also influenced by coexpression with DVL2 (Fig. 3D). The observed differences in the SDS-PAGE migration pattern upon coexpression of the pEGFP::AGS3 constructs with DVL2 are consistent with changes in the phosphorylation status of AGS3. These data suggest that the stabilization of a population of AGS3 into punctate structures containing DVL2 is dependent upon dynamic phosphorylation of the GPR domain of AGS3 (see also Fig. 4D and Discussion).
Fig. 3.
Analysis of AGS3 distribution upon co-expression with DVL2. (A,B) Subcellular distribution of AGS3 in COS-7 cells transfected with pEGFP::AGS3-WT (250 ng), pEGFP::AGS3-Q182H (250 ng), pEGFP::AGS3-T602A (250 ng), pEGFP::AGS3-PM10 (250 ng) and pEGFP::AGS3-PM10-A602 T (250 ng) and/or pRc/CMV2 Dvl-2-Myc (1000 ng). (C) Percentage (mean±s.e.m.) of COS-7 cells with WT AGS3 puncta in the absence of DVL2 co-expression, compared to cells with AGS3-T602A puncta on DVL2 co-expression. 200 cells were counted for each independent transfection. *P<0.05 by two-tailed Student's t-test. (D). Immunoblotting of COS-7 cell lysates of cells expressing WT AGS3 and AGS3-PM constructs with and without co-expression of DVL2. The images shown are representative of three independent experiments. Scale bars: 10 µm.
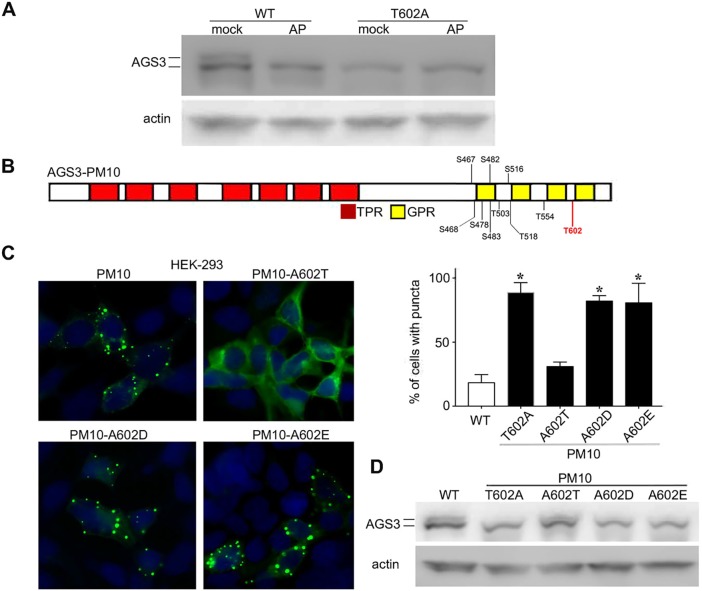
Fig. 4.
Analysis of AGS3 phosphorylation status and subcellular positioning of AGS3. (A) Immunoblotting of cell lysates from COS-7 cells expressing WT AGS3 and AGS3-T602A, with and without alkaline phosphatase treatment. n=5 separate experiments. (B) Schematic representation of serine (S) and threonine (T) residues in the G-protein regulatory domain of AGS3-PM10. (C) Subcellular distribution of AGS3-PM constructs in HEK-293 cells. The alanine residue at 602 in AGS3-PM10 was reversed to threonine as in WT AGS3, or to the acidic amino acids glutamate or aspartate. Right: percentage (mean±s.e.m) of cells with WT AGS3 puncta compared to the percentage of AGS3-PM-expressing cells with cellular puncta. 200 cells were counted for each independent transfection (n=3). *P<0.05 by one-way ANOVA followed by Tukey's multiple comparison test. (D). Immunoblotting of cell lysates generated from the transfection series shown in C.
The role of phosphorylation on the assembly of AGS3 puncta
The relationship between the change in the migration pattern of AGS3-T602A following SDS-PAGE and immunoblotting to actual changes in AGS3 phosphorylation was addressed by treatment of cell extracts with alkaline phosphatase (AP). Incubation of COS-7 cell lysates expressing WT AGS3 with AP, followed by SDS-PAGE and immunoblotting, resulted in a migration pattern of AGS3 that was similar to that observed with AGS3-T602A (Fig. 4A). However, AP treatment of cell lysates expressing AGS3-T602A did not alter the gel migration pattern of the AGS3 phosphomutant. These data are consistent with a role of T602 in AGS3 phosphorylation, either directly or as part of a larger sequential series of serine and threonine phosphorylation events that might be triggered by the initial phosphorylation of T602. The AGS3 sequence was evaluated by PhosphoMotif Finder (http://hprd. org/PhosphoMotif_finder) at the Human Protein Reference Database for candidate phosphorylation sites. As is not uncommon, multiple consensus sites for prediction of serine and threonine phosphorylation were identified. The T602 residue is embedded in a consensus sequence (602TMPD605) for casein kinase II phosphorylation.
As noted above, AGS3-PM10 (Fig. 4B) exhibited a punctate distribution in the cell and this was reversed by conversion of the T602A site mutation back to the WT threonine in the AGS3-PM10-A602T construct. The AGS3-PM10-A602T variant also exhibited a gel migration pattern similar to that of WT AGS3, confirming a key role of T602 as a punctate distribution determinant that likely involves AGS3 phosphorylation (Fig. 4D). We further explored the role of putative T602 phosphorylation in the subcellular distribution of AGS3 by introducing acidic amino acid residues (aspartate, glutamate) at position 602 rather than alanine. AGS3 constructs in which the T602A residue in AGS3-PM10 was substituted with aspartate or glutamate rather than alanine also exhibited the punctate distribution in the cell cytosol. The percentage of cells exhibiting cellular puncta were compared for WT-AGS3 (18.3±3.8%), AGS3-PM10 (88.7±4.5%), AGS3-PM10-A602T (31±2.1%), AGS3-PM10-A602D (82±2.5%) and AGS3-PM10-A602E (80.7±8.8%) (Fig. 4C). Both AGS3-PM10-A602D and AGS3-PM10-A602E also exhibited a gel migration pattern that was similar to that of AGS3-PM10-T602A (Fig. 4D). Often, glutamate and aspartate residues are used as substitutions that might mimic the charge distribution observed upon phosphorylation of serine or threonine. The inability of the aspartate or glutamate substitutions to alter the punctate distribution of AGS3 suggests that a regulatory role of phosphorylation may involve key structural determinants not effectively mimicked by glutamate or aspartate amino acid residues and/or require dynamic phosphorylation and de-phosphorylation as the protein moves among different microenvironments within the cell.
Regulation of AGS3 punctate distribution by stress and G-proteins
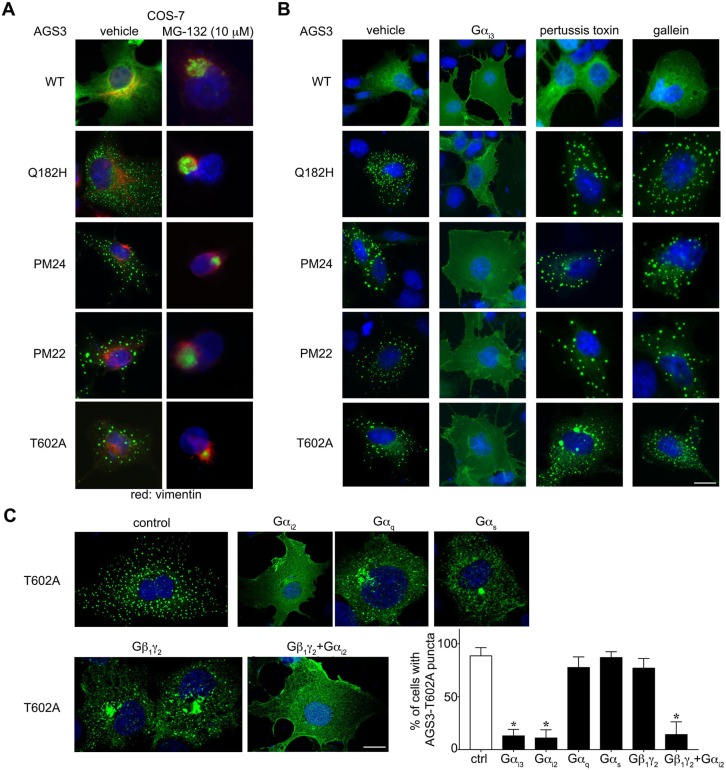
We next asked whether the subcellular distribution of the phosphodeficient variants of AGS3 was similarly regulated by stress and G-protein signaling. The introduction of cellular stress by cell incubation with the proteasome inhibitor MG-132 resulted in disappearance of the cytosolic puncta and the localization of the each of the different constructs (AGS3-Q182H, AGS3-PM24, AGS3-PM22 and AGS3-T602A) to a perinuclear aggresome in coordination with the intermediate filament protein vimentin that typically appears as a ring around the aggresome in this context (Fig. 5A).
Fig. 5.
Regulation of the subcellular distribution of AGS3-PM by cell stress and G-protein signaling. (A,B) Subcellular distribution of AGS3 in COS-7 cells transfected with pEGFP::AGS3-WT (250 ng), pEGFP::AGS3-Q182H (250 ng), pEGFP::AGS3-PM24 (250 ng), pEGFP::AGS3-PM22 (250 ng) and pEGFP::AGS3-T602A (250 ng), with co-expression of pcDNA3::Gαi3 (750 ng) or treatment with MG-132, pertussis toxin or gallein. (C) Subcellular distribution of AGS3-T602A in COS-7 cells transfected with pEGFP::AGS3-T602A (250 ng) alone, and with pcDNA3::Gαi3 (750 ng), pcDNA3::Gαi2 (750 ng), pcDNA3::Gαq (750 ng), pcDNA3::Gαs (750 ng), pcDNA3::Gβ1 (500 ng), and pcDNA3::Gγ2 (500 ng) with and without pcDNA3::Gαi2 (750 ng). Right: percentage (mean±s.e.m) of cells with AGS3-T602A puncta observed with and without co-expression of the indicated G-protein subclasses. 200 cells were counted for each independent transfection (n=3). *P<0.05 by one-way ANOVA followed by Tukey's multiple comparison test. The images shown are representative of five (A,B) or three (C) independent experiments. Scale bars: 10 µm.
AGS3 and other Group II AGS proteins interact with G-protein subunits, and the subcellular distribution and function of Group II AGS proteins may be regulated by various G-protein signaling mechanisms including activation of a G-protein-coupled receptor (Oner et al., 2010). We thus asked whether the subcellular distribution of AGS3-T602A was influenced by G-protein signaling dynamics through expression of G-protein subunits, by uncoupling of G-protein signaling from receptors through cell treatment with pertussis toxin, or by the Gβγ antagonist gallein. As previously noted, the GPR motif in Group II AGS proteins provides a docking site for Gα subunits of the Gαi and Gαo G-protein subgroup. Co-expression of Gαi1, Gαi2, Gαi3 or Gαo stabilized phosphodeficient AGS3 constructs, AGS3-Q182H and WT AGS3 at the cell cortex (Fig. 5B,C; not all Gα subunit data are shown). These data suggest that the absence of candidate phosphorylation sites in the GPR domain or modification of the TPR domain (AGS3-Q182H) do not alter the interaction of G-protein with the GPR domains in AGS3. Interference with G-protein signaling dynamics through treatment with pertussis toxin or the Gβγ antagonist gallein did not alter the subcellular distribution of WT or mutant AGS3 (Fig. 5B). In contrast to the ‘rescue’ of AGS3-T602A from the puncta by co-expression of Gαi or Gαo, co-expression of Gαq or Gαs from distinct G-protein subgroups or Gβ1γ2 did not stabilize the AGS3 variants at the cell cortex, but rather resulted in a redistribution of AGS3 to a perinuclear region that appears to be a component of the Golgi apparatus (Fig. 5C; not all Gα subunit data are shown). The effect of Gβ1γ2 on the subcellular distribution of AGS3-T602A was reversed by expression together with Gαi2, consistent with the expected competitive binding interactions among the three proteins and the subsequent impact of the subcellular distribution of AGS3. The percentage of cells with AGS3-T602A puncta in the absence of any G-protein subunit co-expression (89.5±4.3%) was unchanged compared to AGS3-T602A puncta observed with co-expression of pcDNA3::Gαq (77.7±5.8%), pcDNA3::Gαs (87±3.2%) and pcDNA3::Gβ1γ2 alone (77±5.3%), but was significantly lower in cells expressing pcDNA3::Gαi3 (13±3.6%), pcDNA3::Gαi2 (11±4.5%), and pcDNA3::Gβ1γ2 with pcDNA3::Gαi2 (14.3±6.9%). (Fig. 5C). The perinuclear distribution of AGS3-T602A with co-expression of Gαq, Gαs or Gβ1γ2 is of particular interest given the regulated distribution of AGS3 to this region of the cell and the regulatory role of G-proteins in Golgi function.
DISCUSSION
The regulated distribution of proteins and lipids to and from specific organelles and microdomains within the cell is at the nexus of signaling specificity and basic cell functional processes. The dysregulation of such is causative in various disease processes and the various checkpoints for regulated movement of proteins are a target for therapeutic intervention. Although many compartmental entities within the cell have an organized vesicle texture, other microdomains may be defined visually or biochemically, but the nature of the assembly, disassembly and functional dynamics of such entities are much less well understood. Cytosolic, extranuclear or nuclear puncta, which may vary in size and number within the cell, represent such a compartmental entity and are associated with various aspects of protein and lipid processing, a wide range of functional roles and the adaptation of the cell to external stimuli. Such puncta are involved with various aspects of signal processing cascades and undergo assembly and disassembly in a regulated manner (Boeynaems et al., 2018; Wu and Fuxreiter, 2016; Alberti, 2017a,b). During the course of a broader series of studies to define the biology and functional role of AGS3, it was discovered that the protein moves in a regulated manner between multiple cellular compartments including cytosol, plasma membrane, Golgi membrane, aggresome and cell puncta, with oscillation between a hydrophilic and hydrophobic phase (Vural et al., 2010; Oner et al., 2010, 2013; An et al., 2008). In one of its transit stops, AGS3 localized to cytosolic puncta in a manner that was conformation dependent and regulated by Gαi and other protein-binding partners (Vural et al., 2010). Here, we explored the role of the GPR domain and the role of candidate sites of protein phosphorylation as potential checkpoints for regulated assembly and disassembly of AGS3 puncta. The results indicate that a single amino acid residue (T602) in the GPR domain of AGS3 stabilizes a conformation of AGS3 that is distributed to or retained in cellular puncta in a regulated manner. Overall, the data presented here are consistent with a role of phosphorylation of AGS3 in the regulation of its distribution into punctate structures. The role of the punctate structures in AGS3 functionality will be of particular significance as we move forward with these studies and the identification of a key phosphorylation site as the determinant of puncta distribution provides a strong platform for the next phase of the studies in this regard.
It is likely that the observations with AGS3 have mechanistic and functional resonance with other protein assemblies within the cell as observed with DVL2 puncta. Such defined puncta or ‘protein assemblies’ are distinct from organelles and vesicles with lipid membranes and may vary in size, number and stoichiometry, and exhibit highly dynamic formation and dissociation rates, expanding the functional portfolio of cell biological processes in ways that we are only beginning to understand. The assembly and disassembly of AGS3 in cytosolic, non-nuclear puncta in a regulated manner may be representative of other types of protein assemblies visualized as granules or puncta such as P bodies, stress granules, JUNQ and/or IPOD inclusion bodies or as observed with neurodegeneration or nutrient deprivation. Our data are consistent with the evolving concept that there are distinct, interacting populations of puncta within the cell, as observed by comparative analysis of puncta containing AGS3, AGS3-T602A, AGS3-Q182H and DVL2. Phosphorylation-deficient AGS3 constructs generated cellular puncta that are distinct from those observed with DVL2. In contrast, WT AGS3, which typically has a non-homogeneous, diffuse distribution in the cytosol and which is a phosphorylated protein, was redistributed to DVL2 puncta upon co-expression of the two proteins. This series of observations suggests that the positioning of AGS3 as part of the DVL2 puncta may be phosphorylation dependent. As is the case for AGS3, dishevelled proteins are also associated with multiple cellular and systems functions, of which the molecular basis of the signal processing involved is not fully delineated. The influence of DVL2 on AGS3 redistribution to puncta suggests a potential interaction of the two proteins in signal processing pathways, perhaps in the context of non-canonical WNT signaling pathways involving dishevelled proteins (Schwarz-Romond et al., 2007; Smalley et al., 2005; Bernatík et al., 2014; Greer et al., 2013; González-Sancho et al., 2013; Mlodzik, 2016; Vural et al., 2010). A further mechanistic understanding of the interplay among the puncta populations containing dishevelled proteins and/or AGS3 will be of great interest going forward.
MATERIALS AND METHODS
Materials
MG-132, pertussis toxin, rapamycin and alkaline phosphatase from bovine intestinal mucosa were purchased from Sigma-Aldrich (St Louis, MO, USA). Gallein was purchased from Tocris Biosciences (Bristol, UK). Anti-vimentin antibody (mouse monoclonal RV203, ab8979) was purchased from Abcam (Cambridge, UK). AGS3 antisera were generated by immunization of rabbits with a glutathione S-transferase–AGS3 fusion protein consisting of the AGS3-GPR domain (A461 to S650) (Groves et al., 2010). Anti-Dvl2 antibody (mouse monoclonal 10B5, sc-8026, lot L1714) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All the antibodies were diluted 1:200 for immunocytochemistry and 1:1000 for immunoblotting. pRc/CMV2 Dvl-2-Myc (plasmid 42194) was obtained from Addgene (Cambridge, MA, USA) as deposited by Robert Lefkowitz and Shin-ichi Yanagawa (Lee et al., 1999). All other materials were obtained as previously described (Oner et al., 2013).
Generation of AGS3 phosphomutants
Wild-type (WT) AGS3 rat cDNA encoding 650 amino acids was used as a template to generate the various candidate phosphorylation-deficient AGS3 constructs. PM24 has 24 alanine-substituted serine and threonine residues: S467, S468, S478, S482, S483, T503, T507, S516, T518, S520, T523, S532, S533, S535, S544, S547, T554, S583, S584, T602, S610, S630, S636 and S650 (Fig. 1A). PM22 has 22 alanine-substituted serine and threonine residues: S467, S468, S478, S482, S483, T503, S516, T518, S520, T523, S532, S533, S535, S544, S547, T554, S583, S584, T602, S610, S630 and S636 (Fig. 1A). PM10 and PM13, respectively, have 10 and 13 alanine-substituted serine and threonine residues; AGS3-PM10: S467, S468, S478, S482, S483, S503, T516, T518, T554 and T602; AGS3-PM13a: S467, S468, S478, S482, S483, S516, T518, S554, S583, S584, T602, S610 and S630; AGS3-PM13b: S467, S468, S478, S482, S483, S516, T518, S554, S583, S584, T602, S610 and S650. We generated an additional series of selected alanine-substituted serine and threonine residue AGS3 constructs by site-directed mutagenesis (Fig. 2A): S467A and S468A, S482 and S483A, S532 and S533A, S583A and 584A, S478A, T503A, S516A, T518A, T554A, T602A and S610A. WT AGS3 and the phosphomutant (PM) constructs were generated in pEGFP such that the protein was labeled at the carboxyl terminus with the variant of GFP. AGS3–eGFP exhibits a subcellular distribution and interaction with G-proteins that is similar to WT AGS3 without the eGPF tag. AGS3-GPR region PM plasmids AGS3-PM22 and AGS3-PM10 were generated in the laboratory of D.M. at the University of California, Santa Barbara, CA, USA.
Cell culture, cellular transfection, immunoblotting and fractionation
Cells were cultured in DMEM high-glucose medium supplemented with 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and 5% (HEK-293) or 10% (COS-7, HeLa) fetal bovine serum. All the cell lines are from American Type Culture Collection (Manassas, VA, USA) and they were not recently authenticated and tested for contamination. HEK-293 and COS-7 cells were transfected using 4 μg polyethylenimine (PEI) per ml of cell medium, as described previously (Oner et al., 2013). HeLa cells were transfected as described (Vural et al., 2016). Unless indicated otherwise, cells are typically transfected with 200 ng of plasmid and processed for image analysis and/or immunoblotting after 24 h. In some experiments, 24 h following plasmid transfection, cells were incubated with MG 132 (10 µM), pertussis toxin (200 ng/ml) or gallein (20 µM) for 12 to 18 h prior to harvesting the cells for sample processing.
Cells were lysed with a buffer consisting of 25 mM HEPES (pH 7.4), 4% glycerol, 0.5% (v/v) Nonidet P-40, 150 mM NaCl, 2 mM CaCl2, protease inhibitor cocktail (P8340, Sigma-Aldrich), and a PhosSTOP inhibitor tablet for phosphatase (Roche Applied Sciences, Indianapolis, IN, USA). Lysates were shaken on ice for 15–20 min followed by centrifugation at 10,000 rpm (9279 g) for 10 min at 4°C; 10× protein loading buffer was added to supernatant fraction, which was then and boiled for 10 min. The same lysis buffer without PhosSTOP inhibitor was used to lyse cells that were processed for incubation with alkaline phosphatase. COS-7 cells lysates (60 µg) were incubated with 60 units of alkaline phosphatase for 1 h at 37°C prior to processing for immunoblotting. The fractionation of cellular lysates was as described (Oner et al., 2010; Vural et al., 2010). To allow resolution of multiple AGS3 species by gel electrophoresis, samples were processed by denaturing polyacrylamide gel electrophoresis using Novex 4–20% gradient gels (Thermo Fisher Scientific, Waltham, MA, USA). Fractionated protein samples were run on 10% SDS-PAGE gels. Gel transfers were immunoblotted with anti-actin or anti-tubulin sera for comparative loading controls.
Fluorescence confocal microscopy and image analysis
HEK-293, COS-7 and HeLa cells were processed for fluorescent microscopy and immunofluorescent microscopy as described (Vural et al., 2016; Oner et al., 2013). HEK-293 and COS-7 cell images were captured with a Leica CTR5500 deconvolution fluorescence microscope using a 40× or 63× oil immersion objective as described (Vural et al., 2010). HeLa cell images were captured using PerkinElmer Ultraview spinning-wheel confocal system mounted on a Zeiss Axiovert 200 equipped with an argon-krypton laser with 100× Plan-Aprochromax oil-immersion objective (numerical aperture 1.4) using the image analysis software Volocity 4.1 (PerkinElmer). All images were obtained from approximately the middle plane of the cells. Adobe Photoshop CC 2015.5 was used to process the images. The number of AGS3 puncta and their length was analyzed using ImageJ software (NIH). Briefly, ten different images of cells with eGFP-tagged AGS3 puncta both from COS-7 and HEK-293 cells were converted to 8-bit grayscale and inverted to darken the puncta on a clear background. Following brightness and contrast adjustments, the images were thresholded to 1-bit binary black and white from 8 bit (256 shades of gray) before the enumeration. Puncta were counted by the ‘analyze particles’ option under ‘analyze’ menu. The diameter of the AGS3 puncta in HEK-293 and COS-7 cells was measured by setting the scale of the known distance and measuring the diameter of smallest and biggest puncta in twenty separate COS-7 and HEK-293 cells.
Statistical analysis
Individual COS-7 or HEK-293 cells containing, respectively, >20 cytosolic puncta per cell or >5 cytosolic puncta per cell, following expression of AGS3 constructs, are defined as having a punctate protein distribution, and at least 200 cells were counted in each individual experiment to determine the percentage of cells containing AGS3 puncta. Data are expressed as mean±s.e.m. as determined from at least five independent experiments. Data were analyzed with Prism for Mac OS X (Version 7.0a) software (GraphPad Software, San Diego, CA, USA) using either the two-tailed Student’s t-test or one-way ANOVA, where significant differences between groups were determined by Tukey's multiple comparison test. P-values <0.05 were considered statistically significant.
Acknowledgements
The authors appreciate the valued suggestions and gracious engagement of the many students, fellows, colleagues and collaborators with the body of work involving activators of G-protein signaling. S.M.L. also acknowledges the contributions of key individuals in Turkey that have specifically contributed to the conceptual and technical development of this project with AGS3.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.M.L., A.V., Methodology: S.M.L., A.V., E.F., F.K.; Validation: A.V., E.F., F.K.; Formal analysis: S.M.L., A.V.,; Investigation: S.M.L., A.V., E.F., F.K., D.M.; Resources: S.M.L., D.M.; Data curation: S.M.L., A.V., E.F.; Writing - original draft: S.M.L., A.V.; Writing - review & editing: S.M.L., A.V., E.F., D.M.; Visualization: S.M.L., A.V., E.F.; Supervision: S.M.L., D.M.; Project administration: S.M.L., D.M.; Funding acquisition: S.M.L., D.M.
Funding
This work was supported by National Institutes of Health grants NS24821 and DA025896 to S.M.L. and National Institutes of Health grant 5R21MH086853 to D.M. E.F. was supported by a Visiting Scientist Fellowship grant from the Türkiye Bilimsel ve Teknolojik Araştirma Kurumu. Deposited in PMC for release after 12 months.
References
- Alberti S. (2017a). Phase separation in biology. Curr. Biol. 27, R1097-R1102. 10.1016/j.cub.2017.08.069 [DOI] [PubMed] [Google Scholar]
- Alberti S. (2017b). The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 130, 2789-2796. 10.1242/jcs.200295 [DOI] [PubMed] [Google Scholar]
- An N., Blumer J. B., Bernard M. L. and Lanier S. M. (2008). The PDZ and band 4.1 containing protein Frmpd1 regulates the subcellular location of activator of G-protein signaling 3 and its interaction with G-proteins. J. Biol. Chem. 283, 24718-24728. 10.1074/jbc.M803497200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatík O., Šedová K., Schille C., Ganji R. S., Červenka I., Trantírek L., Schambony A., Zdráhal Z. and Bryja V. (2014). Functional analysis of dishevelled-3 phosphorylation identifies distinct mechanisms driven by casein kinase 1ɛ and frizzled5. J. Biol. Chem. 289, 23520-23533. 10.1074/jbc.M114.590638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer J. B. and Lanier S. M. (2014). Activators of G protein signaling exhibit broad functionality and define a distinct core signaling triad. Mol. Pharmacol. 85, 388-396. 10.1124/mol.113.090068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer J. B., Chandler L. J. and Lanier S. M. (2002). Expression analysis and subcellular distribution of the two G-protein regulators AGS3 and LGN indicate distinct functionality. Localization of LGN to the midbody during cytokinesis. J. Biol. Chem. 277, 15897-15903. 10.1074/jbc.M112185200 [DOI] [PubMed] [Google Scholar]
- Blumer J. B., Bernard M. L., Peterson Y. K., Nezu J., Chung P., Dunican D. J., Knoblich J. A. and Lanier S. M. (2003). Interaction of activator of G-protein signaling 3 (AGS3) with LKB1, a serine/threonine kinase involved in cell polarity and cell cycle progression: phosphorylation of the G-protein regulatory (GPR) motif as a regulatory mechanism for the interaction of GPR motifs with Gi alpha. J. Biol. Chem. 278, 23217-23220. 10.1074/jbc.C200686200 [DOI] [PubMed] [Google Scholar]
- Blumer J. B., Smrcka A. V. and Lanier S. M. (2007). Mechanistic pathways and biological roles for receptor-independent activators of G-protein signaling. Pharmacol. Ther. 113, 488-506. 10.1016/j.pharmthera.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer J. B., Lord K., Saunders T. L., Pacchioni A., Black C., Lazartigues E., Varner K. J., Gettys T. W. and Lanier S. M. (2008). Activator of G protein signaling 3 null mice: I. Unexpected alterations in metabolic and cardiovascular function. Endocrinology 149, 3842-3849. 10.1210/en.2008-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer J. B., Oner S. S. and Lanier S. M. (2012). Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta Physiol. (Oxf.) 204, 202-218. 10.1111/j.1748-1716.2011.02327.x [DOI] [PubMed] [Google Scholar]
- Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., van den Bosch L. et al. (2018). Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420-435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers M. S., Mcfarland K., Lake R. W., Peterson Y. K., Lapish C. C., Gregory M. L., Lanier S. M. and Kalivas P. W. (2004). Activator of G protein signaling 3: a gatekeeper of cocaine sensitization and drug seeking. Neuron 42, 269-281. 10.1016/S0896-6273(04)00159-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branham-O'connor M., Robichaux W. G. III, Zhang X.-K., Cho H., Kehrl J. H., Lanier S. M. and Blumer J. B. (2014). Defective chemokine signal integration in leukocytes lacking activator of G protein signaling 3 (AGS3). J. Biol. Chem. 289, 10738-10747. 10.1074/jbc.M113.515031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishiki K., Kamakura S., Hayase J. and Sumimoto H. (2017). Ric-8a, an activator protein of Galphai, controls mammalian epithelial cell polarity for tight junction assembly and cystogenesis. Genes Cells 22, 293-309. 10.1111/gtc.12477 [DOI] [PubMed] [Google Scholar]
- Choi I. L.-W., Ahn D. W., Choi J.-K., Cha H.-J., Ock M. S., You E. A., Rhee S. M., Kim K. C., Choi Y. H. and Song K. S. (2016). Regulation of airway inflammation by G-protein regulatory motif peptides of AGS3 protein. Sci. Rep. 6, 27054 10.1038/srep27054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukukawa C., Ueda K., Nishidate T., Katagiri T. and Nakamura Y. (2010). Critical roles of LGN/GPSM2 phosphorylation by PBK/TOPK in cell division of breast cancer cells. Genes Chromosomes Cancer 49, 861-872. 10.1002/gcc.20795 [DOI] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ear J., Farquhar M. G. and Ghosh P. (2011). A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol. Biol. Cell 22, 673-686. 10.1091/mbc.e10-08-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Sancho J. M., Greer Y. E., Abrahams C. L., Takigawa Y., Baljinnyam B., Lee K. H., Lee K. S., Rubin J. S. and Brown A. M. C. (2013). Functional consequences of Wnt-induced dishevelled 2 phosphorylation in canonical and noncanonical Wnt signaling. J. Biol. Chem. 288, 9428-9437. 10.1074/jbc.M112.448480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer Y. E., Fields A. P., Brown A. M. C. and Rubin J. S. (2013). Atypical protein kinase Ciota is required for Wnt3a-dependent neurite outgrowth and binds to phosphorylated dishevelled 2. J. Biol. Chem. 288, 9438-9446. 10.1074/jbc.M112.448282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves B., Gong Q., Xu Z., Huntsman C., Nguyen C., Li D. and Ma D. (2007). A specific role of AGS3 in the surface expression of plasma membrane proteins. Proc. Natl. Acad. Sci. USA 104, 18103-18108. 10.1073/pnas.0709282104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves B., Abrahamsen H., Clingan H., Frantz M., Mavor L., Bailey J. and Ma D. (2010). An inhibitory role of the G-protein regulator AGS3 in mTOR-dependent macroautophagy. PLoS ONE 5, e8877 10.1371/journal.pone.0008877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Du Q., Chen X., Zheng Z., Balsbaugh J. L., Maitra S., Shabanowitz J., Hunt D. F. and Macara I. G. (2010). Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. 20, 1809-1818. 10.1016/j.cub.2010.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Pavlov T. S., Nozu K., Rasmussen S. A., Ilatovskaya D. V., LERCH-Gaggl A., North L. M., Kim H., Qian F., Sweeney W. E. Jr. et al. (2012). G-protein signaling modulator 1 deficiency accelerates cystic disease in an orthologous mouse model of autosomal dominant polycystic kidney disease. Proc. Natl. Acad. Sci. USA 109, 21462-21467. 10.1073/pnas.1216830110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-S., Ishimoto A. and Yanagawa S.-I. (1999). Characterization of mouse dishevelled (Dvl) proteins in Wnt/Wingless signaling pathway. J. Biol. Chem. 274, 21464-21470. 10.1074/jbc.274.30.21464 [DOI] [PubMed] [Google Scholar]
- Mauriac S. A., Hien Y. E., Bird J. E., Carvalho S. D.-S., Peyroutou R., Lee S. C., Moreau M. M., Blanc J.-M., Geyser A., Medina C. et al. (2017). Defective Gpsm2/Galphai3 signalling disrupts stereocilia development and growth cone actin dynamics in Chudley-McCullough syndrome. Nat. Commun. 8, 14907 10.1038/ncomms14907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M. (2016). The dishevelled protein family: still rather a mystery after over 20 years of molecular studies. Curr. Top. Dev. Biol. 117, 75-91. 10.1016/bs.ctdb.2015.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadella R., Blumer J. B., Jia G., Kwon M., Akbulut T., Qian F., Sedlic F., Wakatsuki T., Sweeney W. E. Jr, Wilson P. D. et al. (2010). Activator of G protein signaling 3 promotes epithelial cell proliferation in PKD. J. Am. Soc. Nephrol. 21, 1275-1280. 10.1681/ASN.2009121224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oner S. S., An N., Vural A., Breton B., Bouvier M., Blumer J. B. and Lanier S. M. (2010). Regulation of the AGS3.Gαi signaling complex by a seven-transmembrane span receptor. J. Biol. Chem. 285, 33949-33958. 10.1074/jbc.M110.138073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oner S. S., Vural A. and Lanier S. M. (2013). Translocation of activator of G-protein signaling 3 to the Golgi apparatus in response to receptor activation and its effect on the trans-Golgi network. J. Biol. Chem. 288, 24091-24103. 10.1074/jbc.M112.444505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S., DE Vries L., Bauvy C., Chantret I., Cluzeaud F., Ogier-Denis E., Vandewalle A. and Codogno P. (2003). The G-protein regulator AGS3 controls an early event during macroautophagy in human intestinal HT-29 cells. J. Biol. Chem. 278, 20995-21002. 10.1074/jbc.M300917200 [DOI] [PubMed] [Google Scholar]
- Pedram P., Zhai G., Gulliver W., Zhang H. and Sun G. (2017). Two novel candidate genes identified in adults from the Newfoundland population with addictive tendencies towards food. Appetite 115, 71-79. 10.1016/j.appet.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Regner K. R., Nozu K., Lanier S. M., Blumer J. B., Avner E. D., Sweeney W. E. Jr. and Park F. (2011). Loss of activator of G-protein signaling 3 impairs renal tubular regeneration following acute kidney injury in rodents. FASEB J. 25, 1844-1855. 10.1096/fj.10-169797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaux W. G. III, Oner S. S., Lanier S. M. and Blumer J. B. (2015). Direct coupling of a seven-transmembrane-span receptor to a Gαi G-protein regulatory motif complex. Mol. Pharmacol. 88, 231-237. 10.1124/mol.115.097741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaux W. G. III, Branham-O'Connor M., Hwang I.-Y., Vural A., Kehrl J. H. and Blumer J. B. (2017). Regulation of chemokine signal integration by activator of G-protein signaling 4 (AGS4). J. Pharmacol. Exp. Ther. 360, 424-433. 10.1124/jpet.116.238436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadaoui M., Konno D., Loulier K., Goiame R., Jadhav V., Mapelli M., Matsuzaki F. and Morin X. (2017). Loss of the canonical spindle orientation function in the Pins/LGN homolog AGS3. EMBO Rep. 18, 1509-1520. 10.15252/embr.201643048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K. and Tsai L.-H. (2005). G protein βγ subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122, 119-131. 10.1016/j.cell.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T., Metcalfe C. and Bienz M. (2007). Dynamic recruitment of axin by Dishevelled protein assemblies. J. Cell Sci. 120, 2402-2412. 10.1242/jcs.002956 [DOI] [PubMed] [Google Scholar]
- Singh V., Raghuwanshi S. K., Smith N., Rivers E. J. and Richardson R. M. (2014). G Protein-coupled receptor kinase-6 interacts with activator of G protein signaling-3 to regulate CXCR2-mediated cellular functions. J. Immunol. 192, 2186-2194. 10.4049/jimmunol.1301875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley M. J., Signoret N., Robertson D., Tilley A., Hann A., Ewan K., Ding Y., Paterson H. and Dale T. C. (2005). Dishevelled (Dvl-2) activates canonical Wnt signalling in the absence of cytoplasmic puncta. J. Cell Sci. 118, 5279-5289. 10.1242/jcs.02647 [DOI] [PubMed] [Google Scholar]
- Vural A., Oner S., An N., Simon V., Ma D., Blumer J. B. and Lanier S. M. (2010). Distribution of activator of G-protein signaling 3 within the aggresomal pathway: role of specific residues in the tetratricopeptide repeat domain and differential regulation by the AGS3 binding partners Gi(alpha) and mammalian inscuteable. Mol. Cell. Biol. 30, 1528-1540. 10.1128/MCB.01018-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vural A., Mcquiston T. J., Blumer J. B., Park C., Hwang I.-Y., Williams-Bey Y., Shi C.-S., Ma D. Z. and Kehrl J. H. (2013). Normal autophagic activity in macrophages from mice lacking Gαi3, AGS3, or RGS19. PLoS ONE 8, e81886 10.1371/journal.pone.0081886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vural A., AL-Khodor S., Cheung G. Y. C., Shi C.-S., Srinivasan L., Mcquiston T. J., Hwang I.-Y., Yeh A. J., Blumer J. B., Briken V. et al. (2016). Activator of G-protein signaling 3-induced lysosomal biogenesis limits macrophage intracellular bacterial infection. J. Immunol. 196, 846-856. 10.4049/jimmunol.1501595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. and Fuxreiter M. (2016). The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell 165, 1055-1066. 10.1016/j.cell.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Xia B., Gong Q., Bailey J., Groves B., Radeke M., Wood S. A., Szumlinski K. K. and Ma D. (2010). Identification of a deubiquitinating enzyme as a novel AGS3-interacting protein. PLoS ONE 5, e9725 10.1371/journal.pone.0009725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C., Li A., Chuang J.-Z., Saito M., Cáceres A. and Sung C.-H. (2013). IGF-1 activates a cilium-localized noncanonical Gβγ signaling pathway that regulates cell-cycle progression. Dev. Cell 26, 358-368. 10.1016/j.devcel.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]