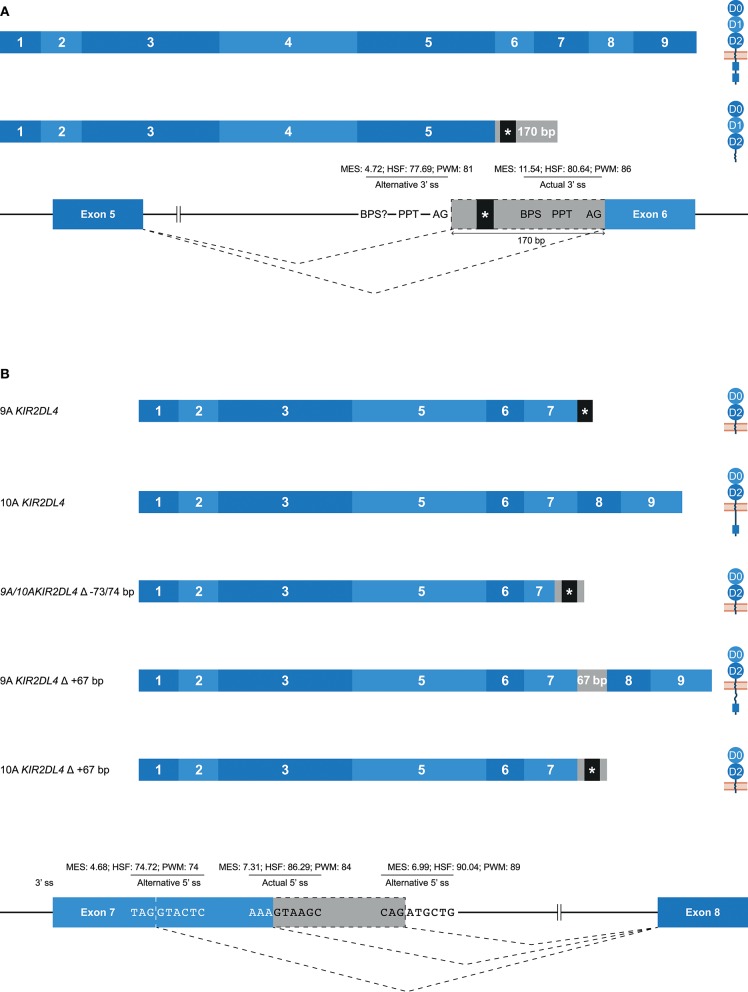
Figure 5.
Alternative splice sites mediate deletions and insertions at the transcription level. The observed transcripts are illustrated, in which exons are indicated by blue boxes, and corresponding protein structures are schematically depicted adjacent to the transcript. Intron inclusions are indicated in gray boxes. Dashed lines indicate the potential splice events, and predicted splicing strength scores are provided for actual and alternative splice sites. Stopcodons are indicated by black boxes with an asterisk (*). (A) The constitutive splicing of human KIR3DL1 results in a transcript including nine exons, and encodes a KIR3DL molecule. Alternative splicing, mediated by an alternative 3′ splice site located 170 bp downstream of the actual splice site, results in a transcript encoding only three extracellular domains. “BPS?” refers to the potential lacking of a BPS for the alternative 3′ ss. (B) The constitutive splicing of human “9A” and “10A” KIR2DL4 alleles results in membrane-bound molecules containing two extracellular domains, or molecules that contain two extracellular domains and a cytoplasmic tail including a single ITIM, respectively. An alternative 5′ ss located in exon 7 results in a partial deletion of the transmembrane region in both “9A” and “10A” KIR2DL4 alleles, and the introduction of a stopcodon. A second alternative 5′ ss located in intron 7 results in a partial intron inclusion of 67 bp. In “9A” KIR2DL4 alleles, this inclusion restores the open reading frame, and these isoforms probably express an inhibitory cytoplasmic tail. In contrast, the same splice event in “10A” KIR2DL4 alleles results in a frame-shift that introduces a stopcodon subsequent to exon 7.