Figure 7.
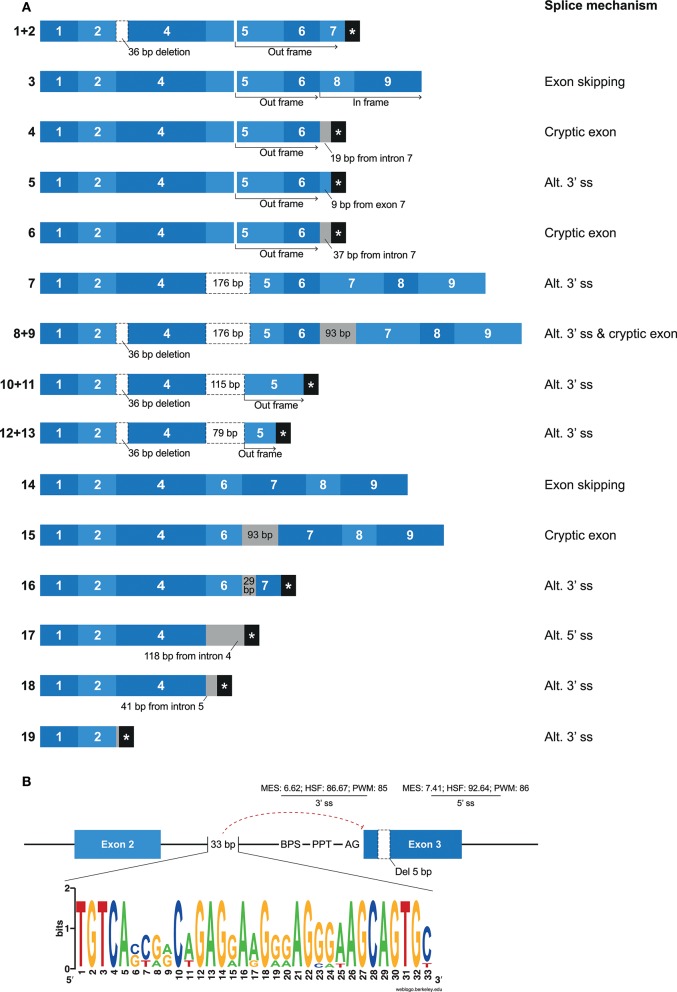
Overview of alternative splicing in Mamu-KIR1D. (A) Nineteen different Mamu-KIR1D transcripts were observed; each transcript is illustrated by blue boxes per exon. White boxes with a dashed outline indicate partial exon deletions, and intronic inclusions are indicated in gray boxes. Multiple transcripts have an out-frame region, due to a deletion of 7 bp in exon 5 at the gDNA level, which can introduce a stopcodon (#1, 2, 4–6). However, the complete inclusion of exon 5 combined with the skipping of exon 7 can restore the open reading frame (#3). The partial deletion of exon 5 might result in Mamu-KIR1D molecules containing an intact inhibitory cytoplasmic tail (#7–9), or in molecules that only consist of a single D1 domain (#10–14). Transcripts that completely skipped exon 5 might encode membrane-bound molecules including an inhibitory cytoplasmic tail (#14, 15), or molecules that only encode the D1 domain, with or without the stem region (#16–18). A second variant was observed for some transcripts, which involved the deletion of 36 bp at the beginning of exon 4, which was also observed in lineage II KIR genes (#1/2, 8/9, 10/11, 12/13; Table 2). (B) Exon 3 is present at the gDNA level in Mamu-KIR1D, but none of the KIR1D isoforms contain the D0 domain encoded by this exon. The BPS, PPT, and both 3′ and 5′ splice sites of exon 3 are intact, just as in human lineage III KIR genes, and predicted splicing strength scores are provided. Intron 2 of all lineage III KIR genes, including Mamu-KIR1D, lack 33 bp in intron 2 compared to all other KIR genes. The lack of this 33 bp stretch might inhibit constitutive splicing of exon 3, as indicated by the red dashed line. The weblogo plot shows the nucleotide sequence composition of this 33 bp stretch that is present in all KIR genes except the KIR lineage III genes, which might mediate the constitutive splicing of exon 3.