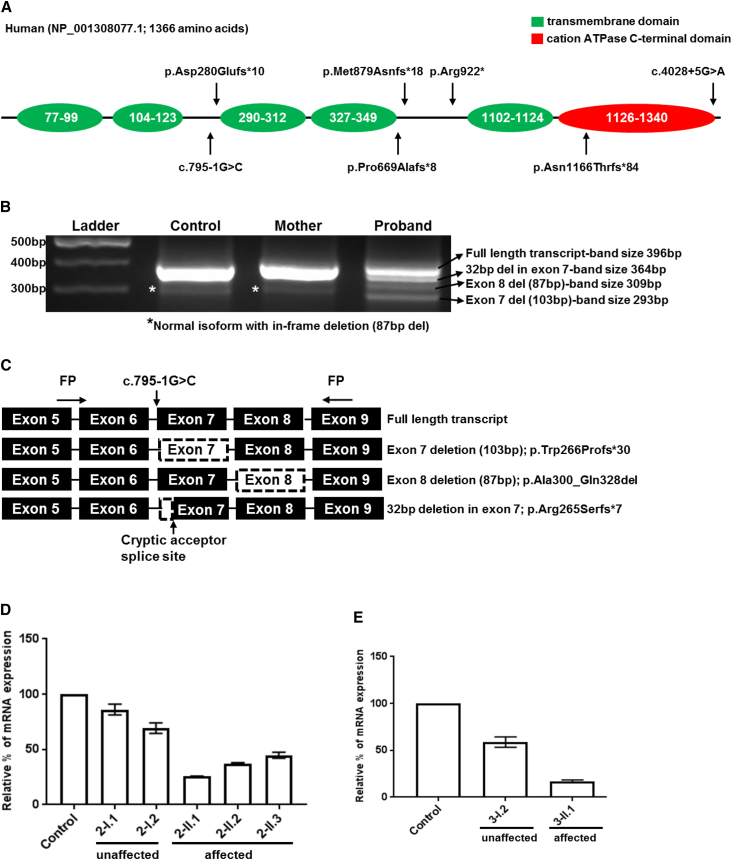
Figure 2.
Molecular Analysis and Gene Expression Studies
(A) Diagrammatic representation of conserved domains of TMEM94 and the position of variants in six families identified.
(B) Splice analysis of the variant NM_001321148.1: c.795-1G>C identified in the proband of family 3. For control and unaffected 20 μL of PCR products were loaded into each well, while for the proband 40 μL was loaded due to severely reduced abundance of transcripts. Normal full-length transcripts (396 bp) and an isoform with exon 8 deletion (309 bp) that predicted to lead to an in-frame deletion(87 bp) was present in control, unaffected and proband. This isoform could be benign. Proband showed two different mutant transcripts; whole exon 7 skipping (p.Trp266Profs∗30) and 32 bp deletion (p.Arg265Serfs∗7) due to the activation of a cryptic acceptor splice site in exon 7.
(C) Diagram showing the primer designing scheme for splice analysis and aberrant transcripts identified. Black boxes representing the coding region with corresponding exon numbers. Boxes with dashed outlines represents the skipped exons/areas of exons identified through TA sub cloning and sequencing (Chromatograms in Figure S3). Variants are named in reference to NM_001321148.1 and NP_001308077.1.
(D and E) Relative quantification of mRNA expression in family 2 and 3 showing markedly reduced expression of TMEM94 in the affected individuals of family 2 (D: 2-II.1, 2-II.2 and 2-II.3) and family 3 (E: 3-II.1). Error bars represent SEM.