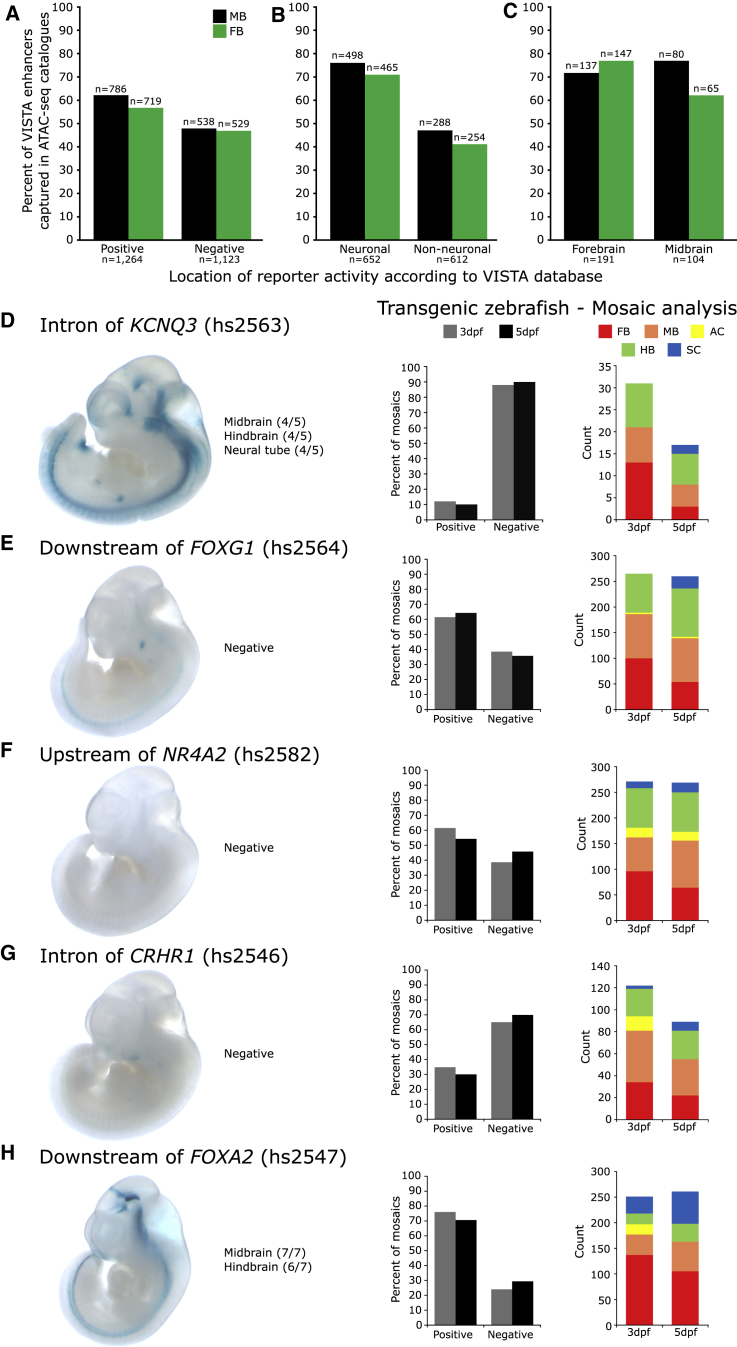
Figure 2.
Validation of the Putative CRE Catalogs In Vivo
(A) Of the elements annotated in VISTA as having enhancer activity, 62% and 56% of these are represented in the MB and FB catalogs, respectively.
(B) An abundance of open-chromatin regions in the MB and FB catalogs overlap confirmed neuronal enhancers (≥70%).
(C) Neuronal enhancers were stratified by the anatomical domains in which they are active; those that are reported active in the MB and FB are enriched in our MB and FB catalogs, respectively.
(D–H) Testing five prioritized putative CREs in vivo identifies five neuronal enhancers.
(D) A putative CRE in intron 1 of KCNQ3 directs expression in the midbrain, hindbrain, and neural tube of E11.5 lacZ reporter mice. It fails to direct expression in a transgenic zebrafish assay at either 3 or 5 days post fertilization (dpf); reporter expression is present in ≤25% of mosaics.
(E, F, and G) Putative CREs downstream of FOXG1, upstream of NR4A2, and in an intron of CRHR1 fail to direct expression in transgenic mice; however, they direct robust neuronal appropriate expression in transgenic zebrafish reporter assays (scored for expression in MB, FB, amacrine cells [ACs], hindbrain [HB], and spinal cord [SC]).
(H) A putative CRE downstream of FOXA2 directs neuronal expression in both transgenic mice and zebrafish assays. n mosaic zebrafish scored: ≥141 for 3 dpf, ≥119 for 5 dpf. All constructs have since been deposited in the VISTA database under the supplied hs numbers.