Figure 3.
Identification of Transcription Factors (TFs) Important to DA Neurons
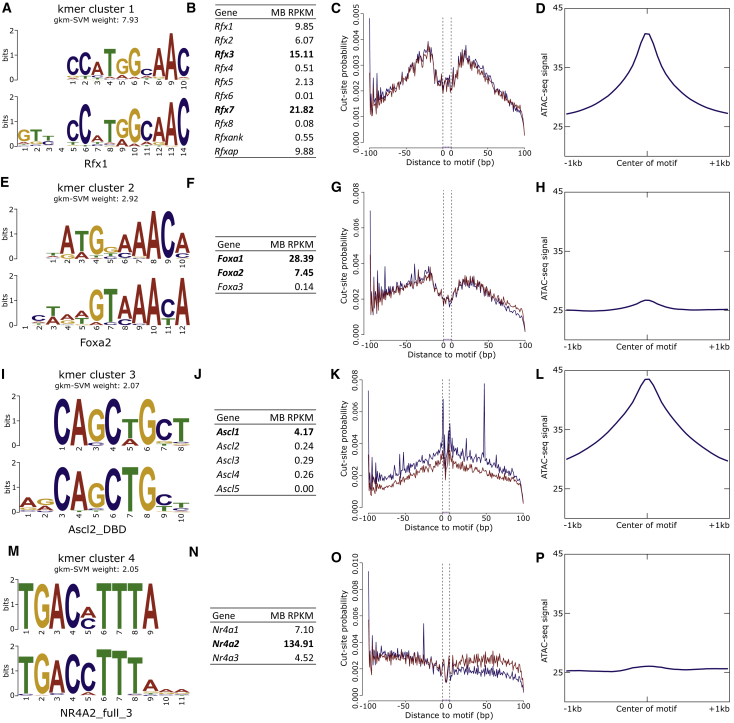
(A) The kmer predicted to have the greatest regulatory potential underlying MB ATAC-seq peaks corresponds to the RFX family of TFs.
(B, C, and D) RNA-seq quantification in these same cells indicates this enrichment is likely due to RFX3 or RFX7 activity. Examining the ATAC-seq signal over predicted binding sites reveals a robust TF footprint (C) and a general enrichment of reads overlapping RFX sites genome-wide (D).
(E–H) Similarly, a kmer corresponding to FOXA1 and/or FOXA2 has similar evidence for the activity of one or both of these TFs.
(I–L) The third-ranked motif most likely corresponds to ASCL1, and although it fails to leave a robust TF footprint (K), there is clear enrichment of ATAC-seq signal overlapping genome-wide predicted ASCL1 binding sites (L).
(M–P) NR4A2, canonically associated with DA neuron biology, is identified as a highly expressed TF that probably contributes to the regulatory potential of the putative CREs; however, it fails to leave a TF footprint in the cut-site patterns around predicted motif sites (O) and is only mildly enriched for ATAC-seq reads over its predicted binding sites (P).