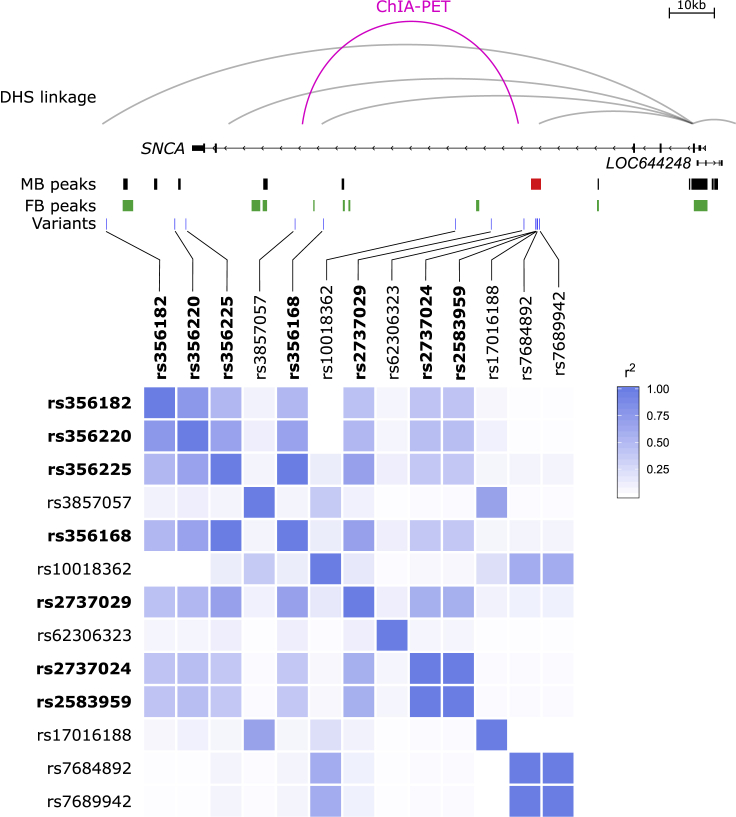
Figure 6.
A Schematic of the Chromatin Interactions, LD Structure, Variation, and Open Chromatin at the SNCA Locus
Publicly available DNase hypersensitivity site (DHS) linkage analysis suggests that the promoter of SNCA possibly interacts with the identified MB-specific enhancer, the lead GWAS variant (dbSNP: rs356182), and a previously functionally validated variant (dbSNP: rs356168). ChIA-PET data suggest that the MB-specific enhancer might interact with variant dbSNP: rs356168. Open-chromatin data from DA neurons do not overlap with any variants at this locus or haplotype other than at the MB-specific enhancer. LD analysis at this locus indicates that despite the low LD structure between the lead GWAS variant (dbSNP: rs356182) and the enhancer-associated variants (dbSNP: rs2737024 and dbSNP: rs2583959), the variants are in the same haplotype. Therefore, the GWAS signal might, at least in part, be flagging the identified enhancer-associated variants.