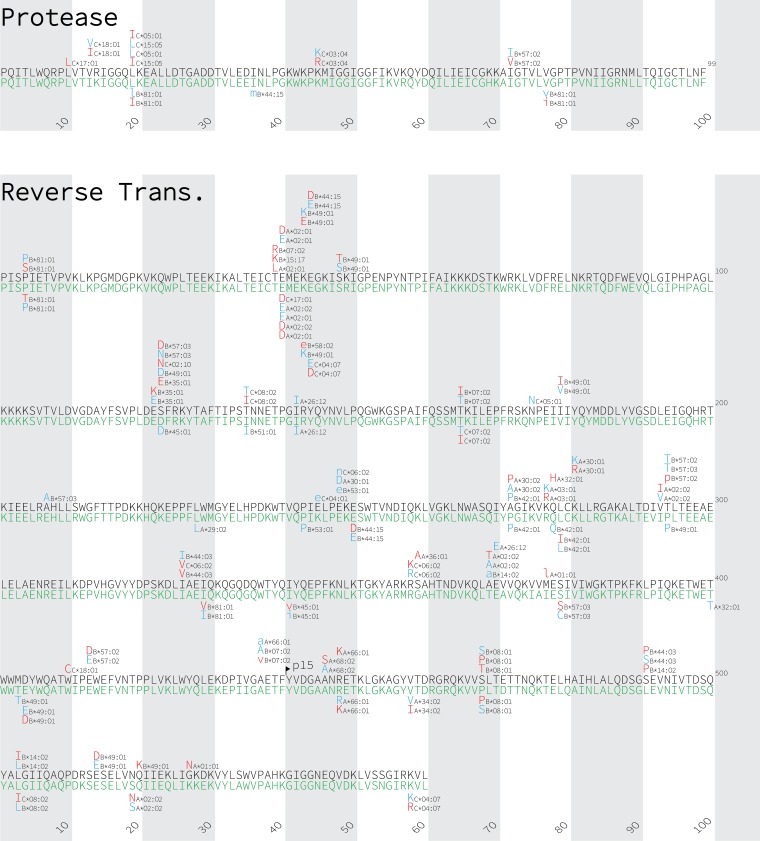
FIG 3.
Gag, Nef, protease, reverse transcriptase, and integrase immune escape maps of HLA-associated polymorphisms identified in subtypes A1 and D. HLA-associated polymorphisms identified in HIV subtype A1 (black consensus sequence) are displayed above; those identified in subtype D (green consensus sequence) are shown below. HIV subtype consensus sequences were obtained from the Los Alamos National Laboratory HIV sequence database, aligned to the HXB2 reference strain, and gap stripped to preserve HXB2 codon numbering. HIV residues that were overrepresented among individuals expressing the given HLA allele are shown in red; these represent the HLA adapted (inferred escaped) form. In contrast, HIV residues that are underrepresented in individuals expressing the given HLA allele are shown in blue; these represent the nonadapted (inferred susceptible) form for that HLA allele. Groups of polymorphisms in close proximity that are selected by the same HLA allele are boxed in yellow. HLA-associated HIV polymorphisms that do not survive correction for HIV codon covariation are shown in lowercase letters. Polymorphisms identified at a q value of <0.2 are shown.