FIG 5.
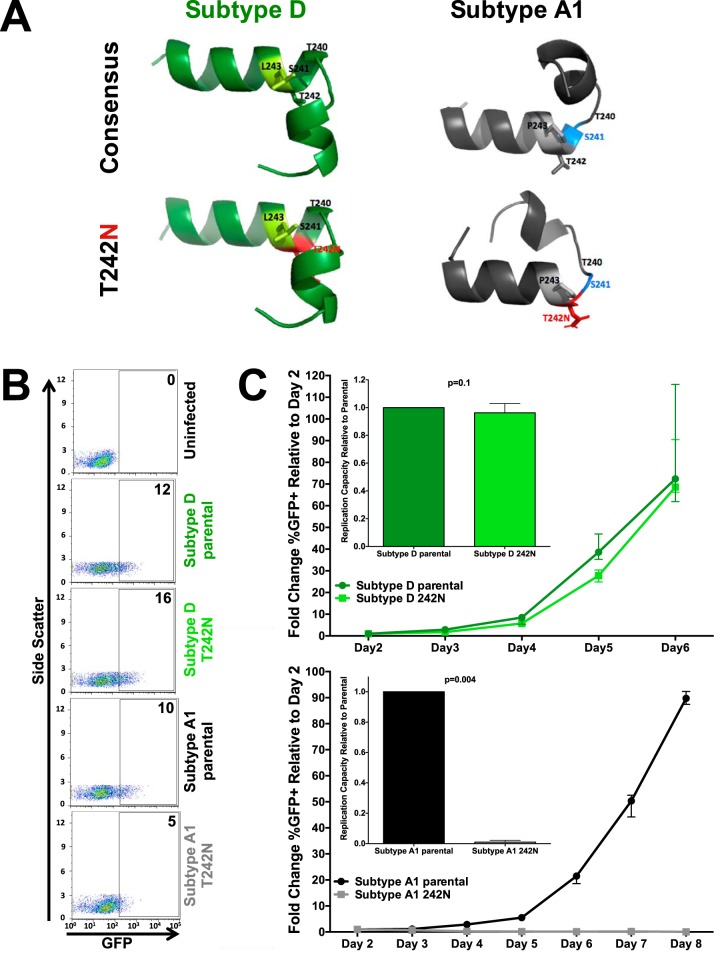
Subtype-specific mutational constraints drive differential B*57:03-mediated selection of Gag T242N between subtypes A1 and D. (A) In silico modeling of the effect of Gag T242N (red, bottom models) on the structure of subtype D (green, left) and A1 (gray, right) consensus p24Gag codons 232 to 254 (top models). Differences in the subtype D and A1 consensus sequence at codon 243 are highlighted (light green, subtype D leucine; light gray, subtype A1 proline). While T242N is predicted to have minimal impact in subtype D (green, left, top versus bottom), its introduction into subtype A1 p24Gag is predicted to substantially alter the peptide structure (gray, right, top versus bottom) by disrupting helix formation (i.e., codon 241, blue) and upstream folding. (B) Representative flow cytometry plots confirming the infectivity (GFP) of VSVg-pseudotyped recombinant virus stocks generated from participant-derived plasma HIV RNA sequences in a CEM-derived LTR-driven GFP reporter T-cell line during experiments to determine the viral titer. Numbers indicate the percentage of GFP-positive (i.e., HIV-infected) cells. (C) Representative curves depicting the rate of spread of VSVg-pseudotyped subtype D (green, top) and subtype A1 (black, bottom) parental (circles; dark green, subtype D; black, subtype A1) and T242N mutant (squares; light green, subtype D; gray, subtype A1) viruses over multiple days in culture. The median and standard error of the percentage of GFP+ (infected) cells relative to that on day 2 are shown. Three independent experiments were performed. (Insets) Replication capacity of parental and T242N mutant viruses (where the value for the latter is normalized to that for the former) calculated over the exponential phase of viral replication. T242N had a modest, though not statistically significant, impact on viral replication in subtype D (median, 0.96 relative to parental subtype D; P = 0.1) but completely abrogated subtype A1 viral replication (P = 0.004).