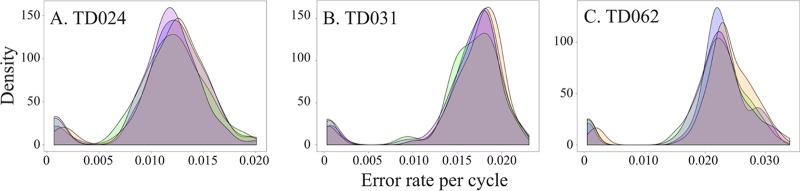
FIG 4.
Error rate per sequencing cycle. Error rates were estimated for each sample using the GATK package using the number of mismatches seen during each cycle and the recalibrated quality score of each position. This gave an estimation of error rate per cycle for each of the 100 sequencing cycles for the first read set of each pair. Error rates per cycle were plotted for samples TD024 (A), TD031 (B), and TD062 (C) for each aligner. Mean error rate per cycle was plotted against the probability density for all 100 cycles. Generally, error rates were consistent over cycles, and there was no evidence of a drop in accuracy over the length of the read. Estimations of mean error rates were a little on the high side (TD024, 1.3%; TD031, 1.7%; TD062, 2.1%), likely caused by the underlying variability of HIV-2. GATK was not able to distinguish between low-frequency variants in the viral population and sequencing errors. However, predicted mean error rates are in line with what would be expected and are informative when choosing a cutoff frequency for reliable SNP calling.