FIGURE 1.
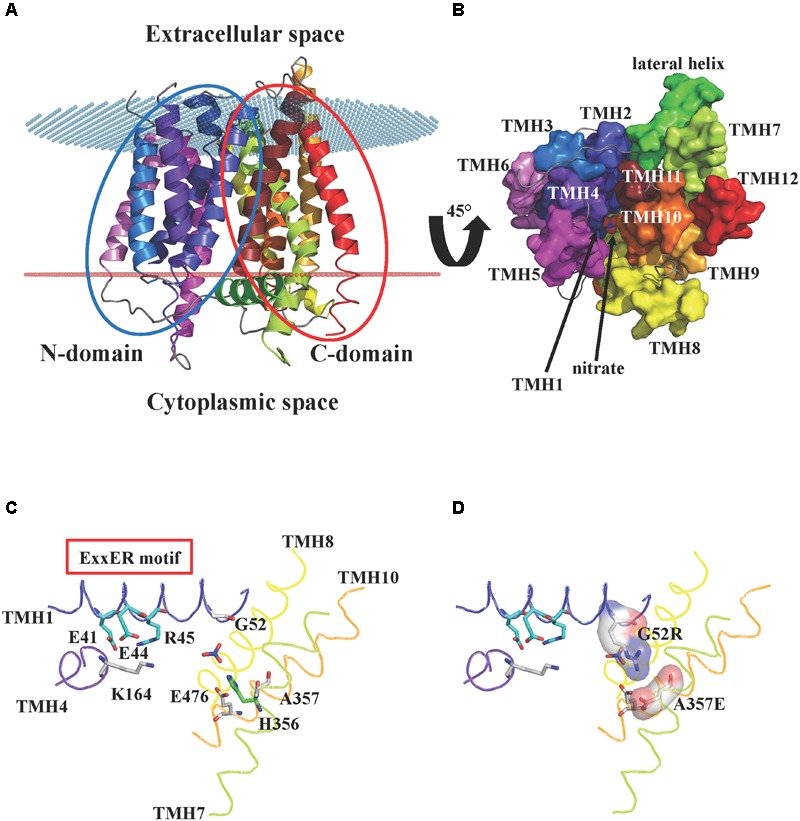
Crystal structure of AtNPF6.3. (A) Cartoon representation of the A. thaliana dual affinity nitrate transporter AtNPF6.3/NRT1.1 (PDB: 4OH3) (Sun et al., 2014) showing the core 12 transmembrane helices (TMHs) arranged into N- and C-terminal bundles (blue and red ovals, respectively). The transporter is in an inward-open conformation with the opening facing the cytoplasm. Spatial position of AtNPF6.3 in the membrane was calculated using the PPM server (Lomize et al., 2012): membrane boundaries are shown as red and blue dots. (B) AtNPF6.3 structure as seen from the cytoplasmic side. Nitrate is located at the bottom of the substrate channel. (C) Close-in view of the substrate channel formed by TMH1, 4, 7, and 10. Important side chains are shown as sticks: Glu41, Glu44 and Arg45 belong to the ExxER/K motif; Lys164 and Glu476 can potentially form an inter-helical salt bridge between TMH4 and TMH10 in the outward open conformation; residues Gly52 and Ala357 correspond to residues that have been shown to form a salt bridge between TMH1 and TMH7 in some bacterial and algal POTs/NRT1s. (D) In silico mutagenesis shows that when residues Gly52 and Ala357 are mutated into arginine and glutamate, respectively, they are within hydrogen bond distance and could form a salt bridge stabilizing the inward-open conformation.
