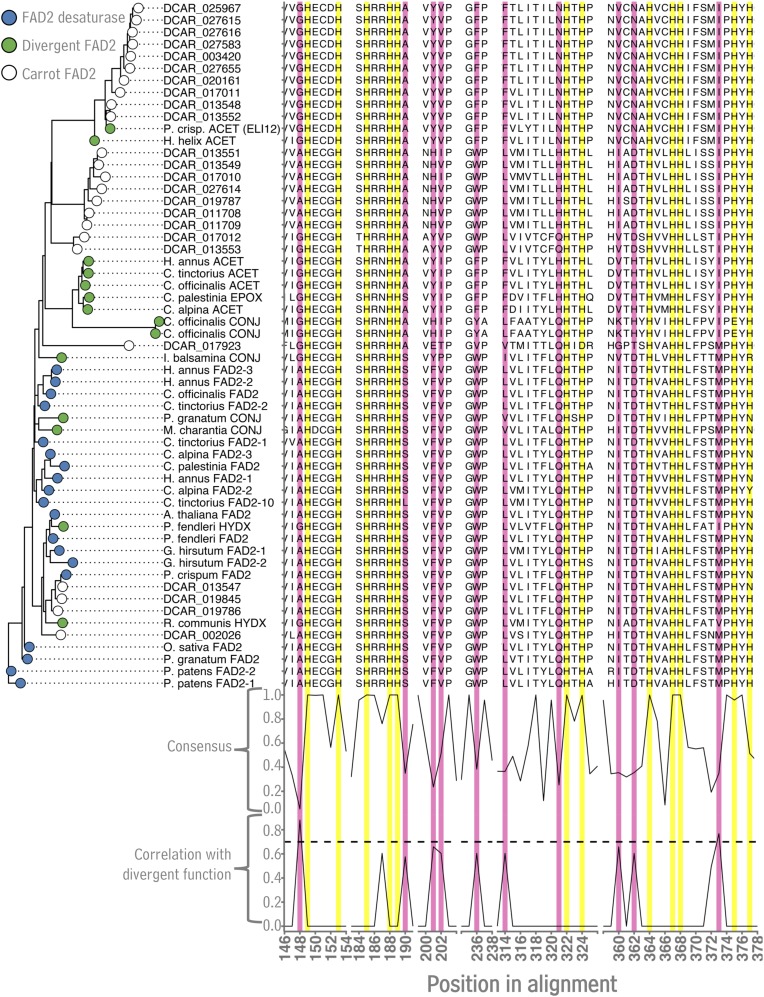
Figure 3.
Analysis of FAD2 amino acid sequences from carrot Daucus carota. Subsets of the multiple sequence alignment of carrot FAD2 sequences and previously characterized FAD2 enzymes from other plant species. Subsets were chosen based on the presence of highly conserved His residues (yellow highlights, “consensus” score on bottom) and residues associated with Δ12 desaturase versus divergent FAD2 activity (pink highlights, “correlation with divergent function” score on bottom). The maximum likelihood phylogenetic tree was generated using nucleotide sequences underlying the amino acids shown in alignment subsets. Sequence data used in this figure can be found in the Phytozome/GenBank databases under the accession numbers listed in Supplemental Table S3.